Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100597 |
| Name | oriT_pL2-87 |
| Organism | Escherichia coli |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_025145 (8019..8104 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | IR1: 7..15, 17..25 (GTCGGGGCT..AGCCCTGAC) IR2: 32..49, 52..69 (GTAATCGTATAGCCGTGC..GCGCGGTTATACGATTAC) |
| Location of nic site | 77..78 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 86 nt
>oriT_pL2-87
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file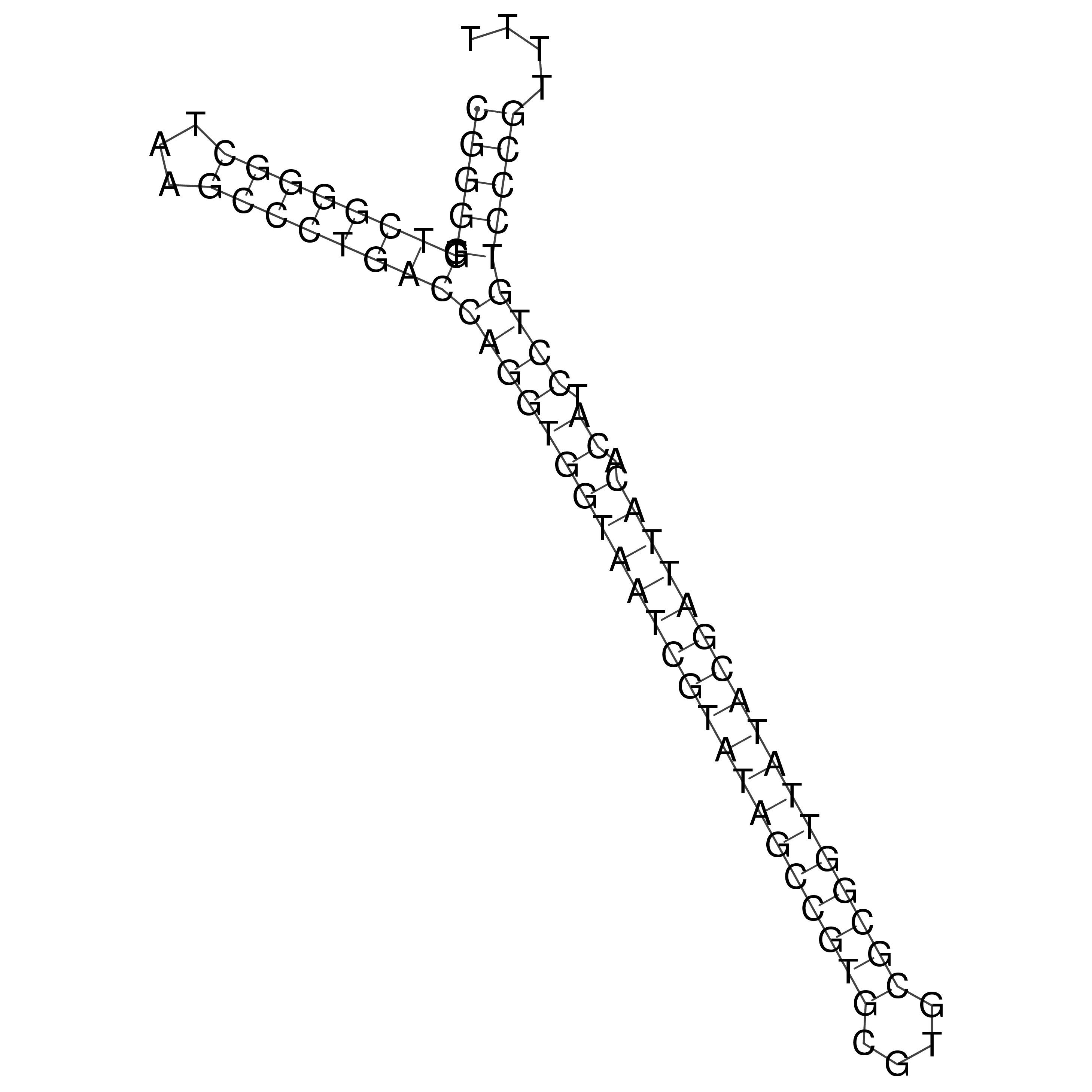
Relaxase
| ID | 669 | GenBank | YP_009069160 |
| Name | NikB_pL2-87 |
UniProt ID | A0A085NWH8 |
| Length | 906 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104812.39 Da Isoelectric Point: 8.3255
>YP_009069160.1 NikB (plasmid) [Escherichia coli]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A085NWH8 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13685..56700
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTF47_RS00080 (D616_p145022) | 11404..13704 | - | 2301 | WP_021553151 | F-type conjugative transfer protein TrbC | - |
| HTF47_RS00085 (D616_p145023) | 13685..14809 | - | 1125 | WP_000152384 | DsbC family protein | trbB |
| HTF47_RS00090 (D616_p145024) | 14806..16113 | - | 1308 | WP_021528768 | IncI1-type conjugal transfer protein TrbA | trbA |
| HTF47_RS00480 | 16650..16745 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HTF47_RS00485 | 17334..17429 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HTF47_RS00100 (D616_p145030) | 17492..17668 | - | 177 | WP_001054907 | hypothetical protein | - |
| HTF47_RS00105 (D616_p145031) | 17819..18286 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| HTF47_RS00110 (D616_p145032) | 18446..19069 | + | 624 | WP_001155250 | helix-turn-helix domain-containing protein | - |
| HTF47_RS00115 (D616_p145033) | 19267..19482 | - | 216 | WP_000266543 | hypothetical protein | - |
| HTF47_RS00120 (D616_p145034) | 19486..19854 | - | 369 | WP_001363143 | hypothetical protein | - |
| HTF47_RS00125 (D616_p145036) | 20154..20396 | - | 243 | WP_001363144 | hypothetical protein | - |
| HTF47_RS00130 | 20726..20878 | + | 153 | WP_001302699 | Hok/Gef family protein | - |
| HTF47_RS00135 (D616_p145038) | 21016..22602 | - | 1587 | WP_000004560 | TnsA endonuclease N-terminal domain-containing protein | - |
| HTF47_RS00140 (D616_p145040) | 22876..23526 | - | 651 | WP_021526891 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HTF47_RS00145 (D616_p145041) | 23597..25762 | - | 2166 | WP_021526892 | DotA/TraY family protein | traY |
| HTF47_RS00150 (D616_p145042) | 25835..26404 | - | 570 | WP_000650755 | conjugal transfer protein TraX | - |
| HTF47_RS00155 (D616_p145043) | 26401..27606 | - | 1206 | WP_001189539 | conjugal transfer protein TraW | traW |
| HTF47_RS00160 (D616_p145044) | 27567..28184 | - | 618 | WP_000286857 | hypothetical protein | traV |
| HTF47_RS00165 (D616_p145045) | 28184..31228 | - | 3045 | WP_021540577 | hypothetical protein | traU |
| HTF47_RS00170 (D616_p145046) | 31453..31998 | + | 546 | WP_001369268 | hypothetical protein | - |
| HTF47_RS00175 (D616_p145047) | 31971..32351 | - | 381 | WP_021540578 | hypothetical protein | - |
| HTF47_RS00180 (D616_p145048) | 32351..32710 | - | 360 | WP_000379509 | hypothetical protein | - |
| HTF47_RS00185 (D616_p145049) | 32769..33395 | - | 627 | WP_001597015 | hypothetical protein | traT |
| HTF47_RS00490 | 33502..33753 | - | 252 | WP_001277575 | hypothetical protein | - |
| HTF47_RS00195 (D616_p145051) | 33810..34208 | - | 399 | WP_000086067 | DUF6750 family protein | traR |
| HTF47_RS00200 (D616_p145052) | 34255..34785 | - | 531 | WP_000987022 | conjugal transfer protein TraQ | traQ |
| HTF47_RS00205 (D616_p145053) | 34782..35495 | - | 714 | WP_000787008 | conjugal transfer protein TraP | traP |
| HTF47_RS00210 (D616_p145054) | 35492..36823 | - | 1332 | WP_009453278 | conjugal transfer protein TraO | traO |
| HTF47_RS00215 (D616_p145055) | 36827..37801 | - | 975 | WP_021532715 | DotH/IcmK family type IV secretion protein | traN |
| HTF47_RS00220 (D616_p145056) | 37812..38507 | - | 696 | WP_001597013 | DotI/IcmL family type IV secretion protein | traM |
| HTF47_RS00225 (D616_p145057) | 38519..38869 | - | 351 | WP_001056369 | hypothetical protein | traL |
| HTF47_RS00230 (D616_p145058) | 38886..42902 | - | 4017 | WP_000842446 | LPD7 domain-containing protein | - |
| HTF47_RS00235 (D616_p145059) | 42966..43256 | - | 291 | WP_000817799 | hypothetical protein | traK |
| HTF47_RS00240 (D616_p145060) | 43253..44401 | - | 1149 | WP_001597012 | plasmid transfer ATPase TraJ | virB11 |
| HTF47_RS00245 (D616_p145061) | 44385..45221 | - | 837 | WP_000848138 | type IV secretory system conjugative DNA transfer family protein | traI |
| HTF47_RS00250 (D616_p145062) | 45218..45670 | - | 453 | WP_001081199 | DotD/TraH family lipoprotein | - |
| HTF47_RS00255 (D616_p145065) | 46029..47231 | - | 1203 | WP_032072259 | conjugal transfer protein TraF | - |
| HTF47_RS00260 (D616_p145066) | 47321..48142 | - | 822 | WP_001597009 | hypothetical protein | traE |
| HTF47_RS00265 (D616_p145068) | 48402..48722 | + | 321 | WP_000695052 | hypothetical protein | - |
| HTF47_RS00270 | 48814..49104 | - | 291 | WP_001597008 | hypothetical protein | - |
| HTF47_RS00275 (D616_p145070) | 49146..50315 | - | 1170 | WP_000486830 | site-specific integrase | - |
| HTF47_RS00280 (D616_p145072) | 50930..52294 | - | 1365 | WP_001597007 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTF47_RS00285 (D616_p145073) | 52299..52934 | - | 636 | WP_000478594 | prepilin peptidase | - |
| HTF47_RS00290 (D616_p145074) | 52953..53426 | - | 474 | WP_000870986 | lytic transglycosylase domain-containing protein | virB1 |
| HTF47_RS00295 (D616_p145075) | 53480..54016 | - | 537 | WP_001011158 | type 4 pilus major pilin | - |
| HTF47_RS00300 (D616_p145076) | 54090..55190 | - | 1101 | WP_000670138 | type II secretion system F family protein | - |
| HTF47_RS00305 (D616_p145077) | 55192..56700 | - | 1509 | WP_000886636 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HTF47_RS00310 (D616_p145078) | 56785..57237 | - | 453 | WP_001112925 | type IV pilus biogenesis protein PilP | - |
| HTF47_RS00315 (D616_p145079) | 57227..58522 | - | 1296 | WP_000129888 | type 4b pilus protein PilO2 | - |
| HTF47_RS00320 (D616_p145080) | 58543..60162 | - | 1620 | WP_001034998 | PilN family type IVB pilus formation outer membrane protein | - |
| HTF47_RS00325 (D616_p145081) | 60193..60630 | - | 438 | WP_000539685 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 1057 | GenBank | NC_025145 |
| Plasmid name | pL2-87 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 87042 bp | Coordinate of oriT [Strand] | 8019..8104 [-] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |