Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100579 |
| Name | oriT_pACM1 |
| Organism | Klebsiella oxytoca |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_024997 (64881..64986 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 18..35, 41..58 (GTTTTTGGAGTACCGCCG..CGGCGGCCGTCCAAAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 106 nt
>oriT_pACM1
AGATAGCTAACCTCGTTAGGGGGTGTCGGGGCTTGCCCTGACCAAGACGTTTTTGGACGGCCGCCGCGTGTCGGCGGTACTCCAAAAACACATCTTGTCCCGTACT
AGATAGCTAACCTCGTTAGGGGGTGTCGGGGCTTGCCCTGACCAAGACGTTTTTGGACGGCCGCCGCGTGTCGGCGGTACTCCAAAAACACATCTTGTCCCGTACT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file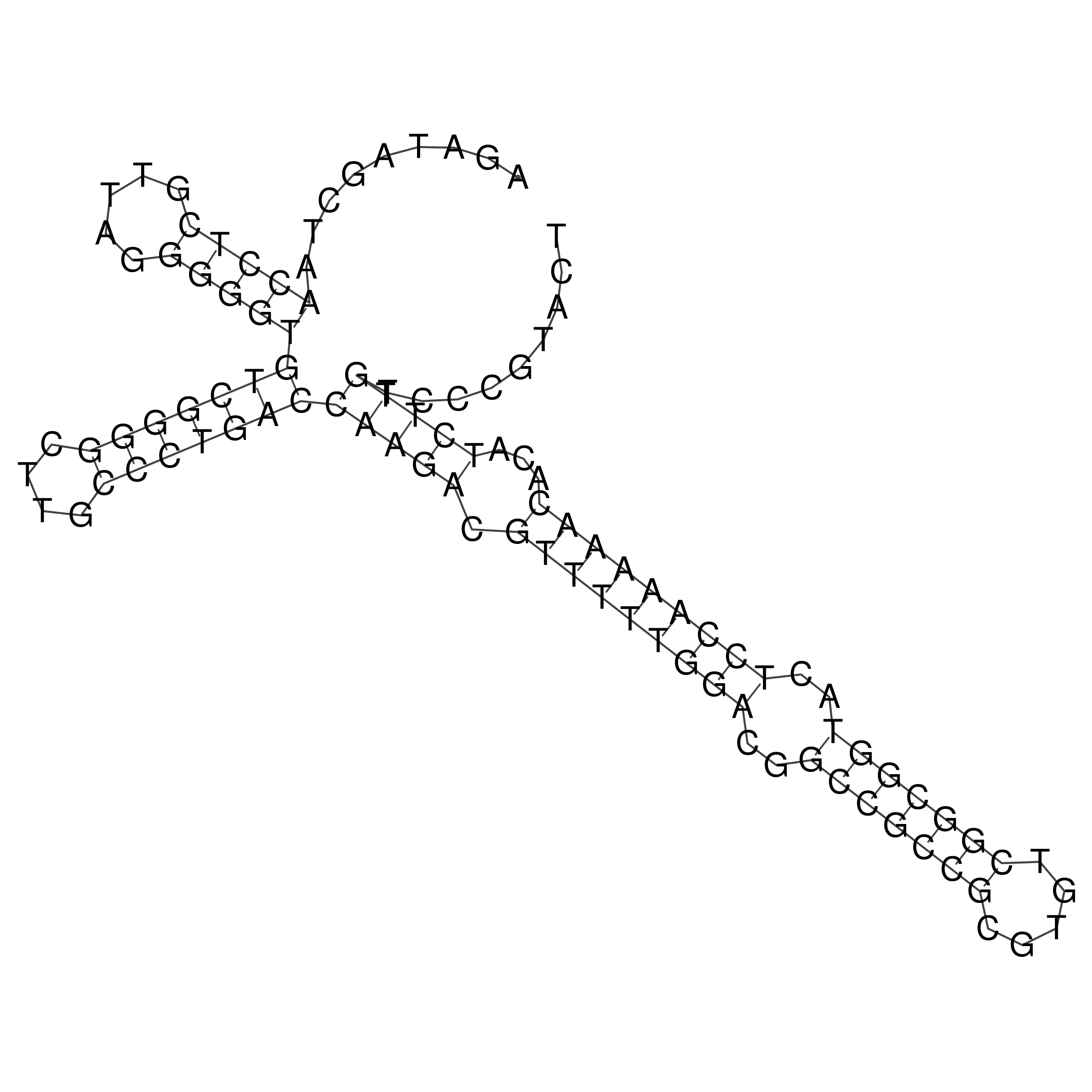
Relaxase
| ID | 651 | GenBank | YP_009062617 |
| Name | PutativerelaxaseofpACM1 |
UniProt ID | A0A088FMU5 |
| Length | 658 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 658 a.a. Molecular weight: 75094.79 Da Isoelectric Point: 9.9948
>YP_009062617.1 putative mobility protein (plasmid) [Klebsiella oxytoca]
MIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVDV
EPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQIFDSVKFTLAS
MGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVNDR
QQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHKT
FADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTAY
TPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSVS
DPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSSR
KKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLTE
SNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQNE
KHRLIREQNNSKNEMQFRSESDDKFNPK
MIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVDV
EPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQIFDSVKFTLAS
MGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVNDR
QQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHKT
FADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTAY
TPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSVS
DPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSSR
KKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLTE
SNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQNE
KHRLIREQNNSKNEMQFRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A088FMU5 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 34839..62085
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTD00_RS00215 (D733_p1038) | 30531..30743 | + | 213 | Protein_41 | IS6 family transposase | - |
| HTD00_RS00220 (D733_p1039) | 30766..31653 | + | 888 | Protein_42 | methyl-accepting chemotaxis protein | - |
| HTD00_RS00225 (D733_p1040) | 31842..32648 | - | 807 | WP_269467505 | Y-family DNA polymerase | - |
| HTD00_RS00230 (D733_p1041) | 32636..33070 | - | 435 | WP_015062799 | translesion error-prone DNA polymerase V autoproteolytic subunit | - |
| HTD00_RS00235 (D733_p1042) | 33223..33876 | - | 654 | WP_031942486 | type II CAAX endopeptidase family protein | - |
| HTD00_RS00240 (D733_p1043) | 33944..34339 | + | 396 | WP_031942487 | DUF1496 domain-containing protein | - |
| HTD00_RS00245 (D733_p1044) | 34444..34839 | + | 396 | WP_004187436 | lytic transglycosylase domain-containing protein | - |
| HTD00_RS00250 (D733_p1045) | 34839..36146 | + | 1308 | WP_015062796 | hypothetical protein | trbA |
| HTD00_RS00255 (D733_p1046) | 36157..37107 | + | 951 | WP_004206886 | DsbC family protein | trbB |
| HTD00_RS00260 (D733_p1047) | 37120..39207 | + | 2088 | WP_004187443 | hypothetical protein | - |
| HTD00_RS00630 | 39258..39353 | - | 96 | WP_004206884 | DinQ-like type I toxin DqlB | - |
| HTD00_RS00265 | 40208..40450 | - | 243 | WP_172686030 | hypothetical protein | - |
| HTD00_RS00270 (D733_p1048) | 40580..41647 | - | 1068 | WP_019725043 | plasmid replication initiator RepA | - |
| HTD00_RS00275 (D733_p1050) | 41932..42162 | - | 231 | WP_004187496 | IncL/M type plasmid replication protein RepC | - |
| HTD00_RS00280 (D733_p1051) | 42236..42889 | - | 654 | WP_004206935 | hypothetical protein | - |
| HTD00_RS00285 (D733_p1052) | 42892..45072 | - | 2181 | WP_004187492 | DotA/TraY family protein | traY |
| HTD00_RS00290 (D733_p1053) | 45065..45715 | - | 651 | WP_004187488 | hypothetical protein | - |
| HTD00_RS00295 (D733_p1054) | 45712..46920 | - | 1209 | WP_011091082 | conjugal transfer protein TraW | traW |
| HTD00_RS00300 (D733_p1055) | 46917..49967 | - | 3051 | WP_031942490 | plasmid transfer protein | traU |
| HTD00_RS00305 | 49964..50458 | - | 495 | WP_011091080 | hypothetical protein | - |
| HTD00_RS00310 (D733_p1056) | 50504..50893 | - | 390 | WP_011091078 | DUF6750 family protein | traR |
| HTD00_RS00315 (D733_p1057) | 50910..51440 | - | 531 | WP_011091077 | conjugal transfer protein TraQ | traQ |
| HTD00_RS00320 (D733_p1058) | 51464..52168 | - | 705 | WP_031942491 | conjugal transfer protein TraP | traP |
| HTD00_RS00325 (D733_p1059) | 52180..53529 | - | 1350 | WP_011091075 | conjugal transfer protein TraO | traO |
| HTD00_RS00330 (D733_p1060) | 53541..54692 | - | 1152 | WP_011091074 | DotH/IcmK family type IV secretion protein | traN |
| HTD00_RS00335 (D733_p1061) | 54701..55483 | - | 783 | WP_011091073 | DotI/IcmL family type IV secretion protein | traM |
| HTD00_RS00340 | 55482..55892 | + | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| HTD00_RS00345 | 55879..56076 | + | 198 | WP_004187464 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| HTD00_RS00350 (D733_p1063) | 56077..56631 | - | 555 | WP_011154467 | hypothetical protein | traL |
| HTD00_RS00610 | 56555..59857 | - | 3303 | WP_031942492 | LPD7 domain-containing protein | - |
| HTD00_RS00360 | 59882..60142 | - | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| HTD00_RS00365 (D733_p1065) | 60132..61295 | - | 1164 | WP_004187313 | plasmid transfer ATPase TraJ | virB11 |
| HTD00_RS00370 (D733_p1066) | 61306..62085 | - | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| HTD00_RS00375 (D733_p1067) | 62082..62582 | - | 501 | WP_004187320 | DotD/TraH family lipoprotein | - |
| HTD00_RS00380 (D733_p1068) | 62596..64575 | - | 1980 | WP_059509498 | TraI/MobA(P) family conjugative relaxase | - |
| HTD00_RS00615 | 64562..65125 | - | 564 | WP_227992076 | plasmid mobilization protein MobA | - |
| HTD00_RS00390 (D733_p1070) | 65124..65519 | + | 396 | WP_031942494 | hypothetical protein | - |
| HTD00_RS00395 (D733_p1071) | 66020..66556 | - | 537 | WP_011091066 | hypothetical protein | - |
| HTD00_RS00400 | 66689..67000 | - | 312 | WP_004187333 | hypothetical protein | - |
Host bacterium
| ID | 1039 | GenBank | NC_024997 |
| Plasmid name | pACM1 | Incompatibility group | IncL/M |
| Plasmid size | 89977 bp | Coordinate of oriT [Strand] | 64881..64986 [+] |
| Host baterium | Klebsiella oxytoca |
Cargo genes
| Drug resistance gene | tet(A), sul1, qacE, ant(3'')-Ia, aac(3)-Ia, dfrA1, blaSHV-5 |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |