Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100534 |
| Name | oriT_pCFSAN002069_01 |
| Organism | Salmonella enterica subsp. enterica serovar Heidelberg str. CFSAN002069 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_021813 (44500..44602 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | IR1: 19..26, 30..37 (GTCGGGGC..GCCCTGAC) IR2: 44..58, 67..81 (GCAATTGTATAATCG..CGGTTATACAATTGC) |
| Location of nic site | 89..90 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 103 nt
>oriT_pCFSAN002069_01
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file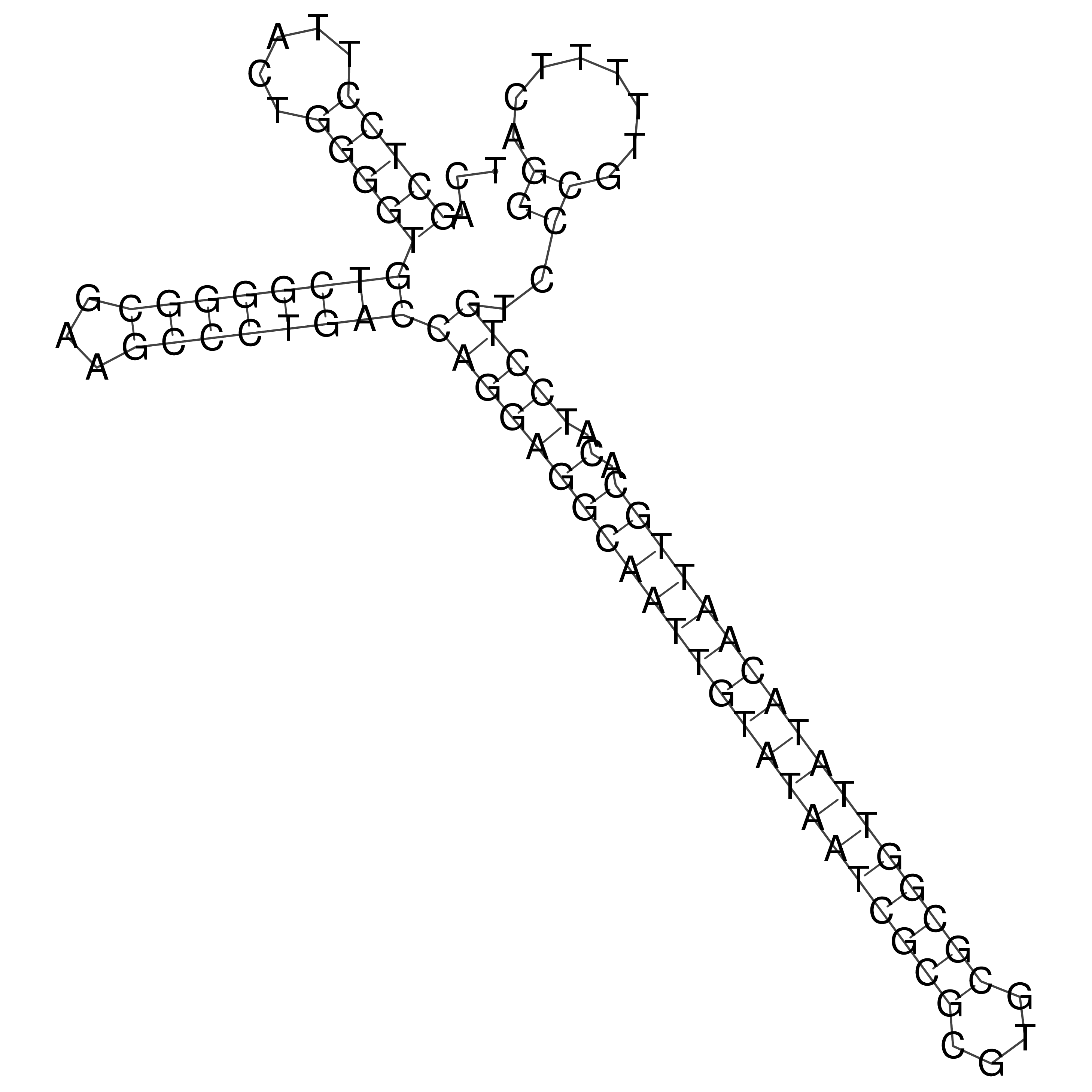
Relaxase
| ID | 605 | GenBank | WP_001354015 |
| Name | NikB_pCFSAN002069_01 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104018.47 Da Isoelectric Point: 7.3526
>WP_001354015.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 835..39134
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CFSAN002069_RS00010 (CFSAN002069_23055) | 372..824 | + | 453 | WP_001247336 | type IV pilus biogenesis protein PilP | - |
| CFSAN002069_RS00015 (CFSAN002069_23050) | 835..2388 | + | 1554 | WP_000362202 | ATPase, T2SS/T4P/T4SS family | virB11 |
| CFSAN002069_RS00020 (CFSAN002069_23840) | 2401..3498 | + | 1098 | Protein_3 | type II secretion system F family protein | - |
| CFSAN002069_RS00025 (CFSAN002069_23040) | 3516..4130 | + | 615 | WP_000959785 | type 4 pilus major pilin | - |
| CFSAN002069_RS00030 (CFSAN002069_23035) | 4140..4700 | + | 561 | WP_000014116 | lytic transglycosylase domain-containing protein | virB1 |
| CFSAN002069_RS23935 | 4685..5341 | + | 657 | WP_001193553 | prepilin peptidase | - |
| CFSAN002069_RS00035 (CFSAN002069_23850) | 5341..6633 | + | 1293 | WP_001417545 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| CFSAN002069_RS26085 | 6630..6878 | - | 249 | WP_001349157 | hypothetical protein | - |
| CFSAN002069_RS26645 | 8116..8400 | - | 285 | WP_187642572 | pilus assembly protein PilV | - |
| CFSAN002069_RS00045 (CFSAN002069_23600) | 8399..9553 | + | 1155 | WP_001139957 | site-specific integrase | - |
| CFSAN002069_RS00050 (CFSAN002069_23595) | 9704..10528 | + | 825 | WP_001238939 | conjugal transfer protein TraE | traE |
| CFSAN002069_RS00055 (CFSAN002069_23590) | 10613..11815 | + | 1203 | WP_000976351 | conjugal transfer protein TraF | - |
| CFSAN002069_RS00060 (CFSAN002069_23585) | 11875..12459 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| CFSAN002069_RS00065 (CFSAN002069_23580) | 12854..13312 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| CFSAN002069_RS00070 (CFSAN002069_23575) | 13309..14127 | + | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| CFSAN002069_RS00075 (CFSAN002069_23570) | 14124..15272 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| CFSAN002069_RS23970 | 15269..15559 | + | 291 | WP_001299214 | hypothetical protein | traK |
| CFSAN002069_RS00080 (CFSAN002069_23560) | 15574..16125 | + | 552 | WP_000014583 | phospholipase D family protein | - |
| CFSAN002069_RS00085 (CFSAN002069_23555) | 16215..19982 | + | 3768 | WP_001141529 | LPD7 domain-containing protein | - |
| CFSAN002069_RS00090 (CFSAN002069_23550) | 20000..20347 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| CFSAN002069_RS00095 (CFSAN002069_23545) | 20344..21036 | + | 693 | WP_000138548 | DotI/IcmL family type IV secretion protein | traM |
| CFSAN002069_RS00100 (CFSAN002069_23540) | 21047..22030 | + | 984 | WP_001191877 | IncI1-type conjugal transfer protein TraN | traN |
| CFSAN002069_RS00105 (CFSAN002069_23535) | 22033..23322 | + | 1290 | WP_001272005 | conjugal transfer protein TraO | traO |
| CFSAN002069_RS00110 | 23322..24026 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| CFSAN002069_RS00115 (CFSAN002069_23525) | 24026..24553 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| CFSAN002069_RS00120 (CFSAN002069_23520) | 24604..25008 | + | 405 | WP_000086957 | IncI1-type conjugal transfer protein TraR | traR |
| CFSAN002069_RS00125 (CFSAN002069_23515) | 25072..25260 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| CFSAN002069_RS00130 (CFSAN002069_23510) | 25244..26044 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| CFSAN002069_RS00135 (CFSAN002069_23505) | 26134..29178 | + | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| CFSAN002069_RS23975 | 29178..29792 | + | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| CFSAN002069_RS00140 (CFSAN002069_23500) | 29759..30961 | + | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| CFSAN002069_RS00145 (CFSAN002069_23495) | 30990..31574 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| CFSAN002069_RS00150 (CFSAN002069_23490) | 31671..33833 | + | 2163 | WP_000698351 | DotA/TraY family protein | traY |
| CFSAN002069_RS00155 (CFSAN002069_23485) | 33898..34560 | + | 663 | WP_000653334 | plasmid IncI1-type surface exclusion protein ExcA | - |
| CFSAN002069_RS00160 (CFSAN002069_23480) | 34632..34841 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| CFSAN002069_RS26530 (CFSAN002069_23475) | 35233..35409 | + | 177 | WP_001054904 | hypothetical protein | - |
| CFSAN002069_RS26915 | 35474..35569 | - | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| CFSAN002069_RS26920 (CFSAN002069_23465) | 36130..36321 | + | 192 | WP_174715448 | hypothetical protein | - |
| CFSAN002069_RS00170 (CFSAN002069_23460) | 36393..36545 | - | 153 | WP_001331364 | Hok/Gef family protein | - |
| CFSAN002069_RS00175 (CFSAN002069_23455) | 36837..38045 | + | 1209 | WP_000121274 | IncI1-type conjugal transfer protein TrbA | trbA |
| CFSAN002069_RS00180 (CFSAN002069_23450) | 38064..39134 | + | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| CFSAN002069_RS00185 (CFSAN002069_23445) | 39127..41418 | + | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 995 | GenBank | NC_021813 |
| Plasmid name | pCFSAN002069_01 | Incompatibility group | IncI1 |
| Plasmid size | 110363 bp | Coordinate of oriT [Strand] | 44500..44602 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Heidelberg str. CFSAN002069 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | copper resistance |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |