Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100527 |
| Name | oriT_pSA8589 |
| Organism | Staphylococcus aureus |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_021230 (3535..3575 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 2..8, 13..19 (ACTTTAT..ATAAAGT) |
| Location of nic site | 27..28 |
| Conserved sequence flanking the nic site |
GTG|TTA |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 41 nt
>oriT_pSA8589
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file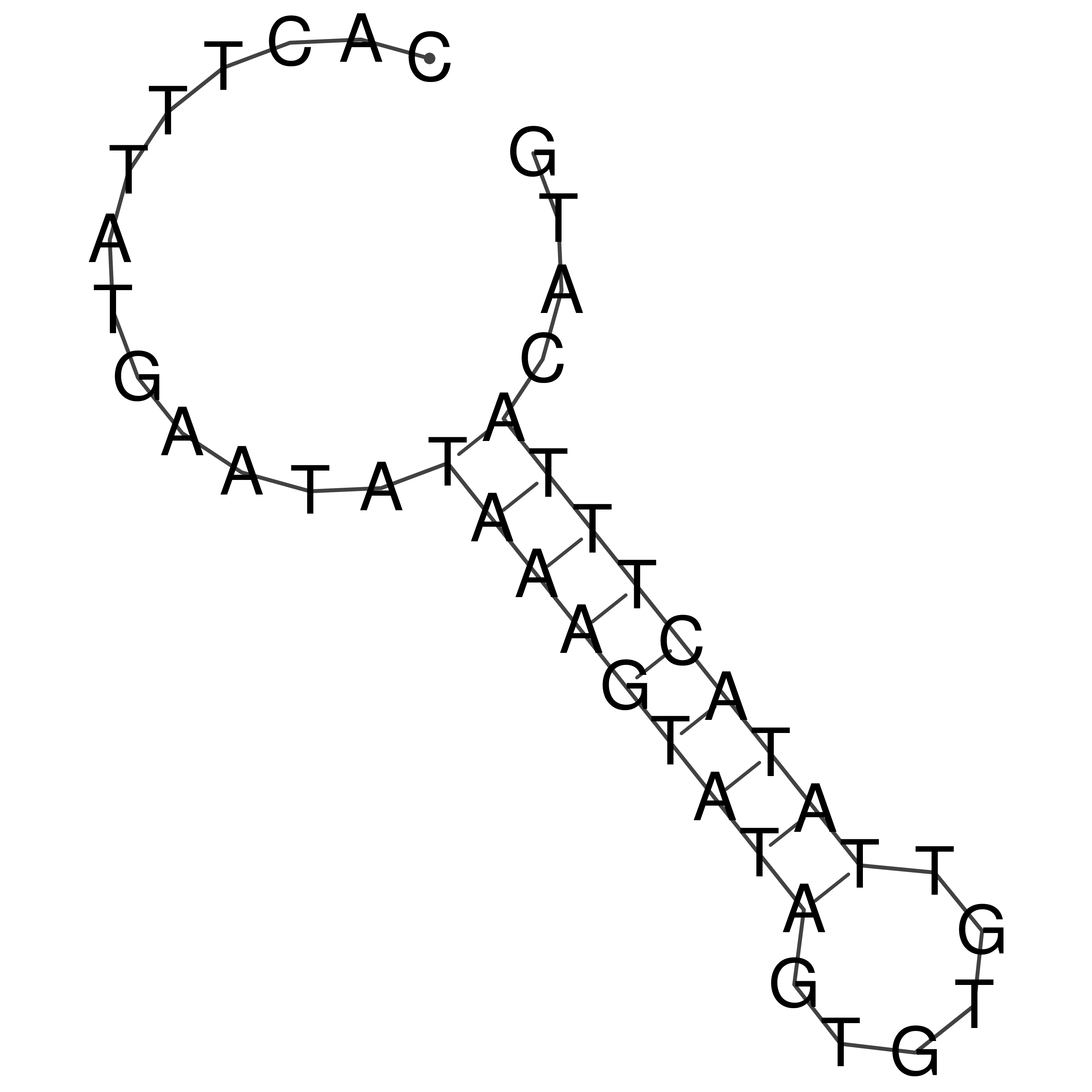
Relaxase
| ID | 598 | GenBank | YP_007988854 |
| Name | Mob_pSA8589 |
UniProt ID | R4NGV8 |
| Length | 408 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 408 a.a. Molecular weight: 48229.55 Da Isoelectric Point: 7.0097
>YP_007988854.1 recombination/mobilization protein (plasmid) [Staphylococcus aureus]
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKDQIDKNDKDIDDYNDLVKRYNNLYEENTSLKSQIGDLKEEIKLIY
QSTKRFLKDRISDFKAFKEVFKELADNISNISREKGLDSSFKKEFDRENKKKQTRGIR
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKDQIDKNDKDIDDYNDLVKRYNNLYEENTSLKSQIGDLKEEIKLIY
QSTKRFLKDRISDFKAFKEVFKELADNISNISREKGLDSSFKKEFDRENKKKQTRGIR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | R4NGV8 |
Host bacterium
| ID | 988 | GenBank | NC_021230 |
| Plasmid name | pSA8589 | Incompatibility group | _ |
| Plasmid size | 6962 bp | Coordinate of oriT [Strand] | 3535..3575 [+] |
| Host baterium | Staphylococcus aureus |
Cargo genes
| Drug resistance gene | cfr |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |