Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100495 |
| Name | oriT_pB76-81 |
| Organism | Pseudomonas syringae |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_019328 (783..928 [+], 146 nt) |
| oriT length | 146 nt |
| IRs (inverted repeats) | IR1: 38..43, 52..57 (AAAAAG..CTTTTT) IR2: 72..83, 87..98 (GTGCCAAAGGGG..CCCCTTTGAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 146 nt
>oriT_pB76-81
CGAATTCGATAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCAT
CGAATTCGATAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file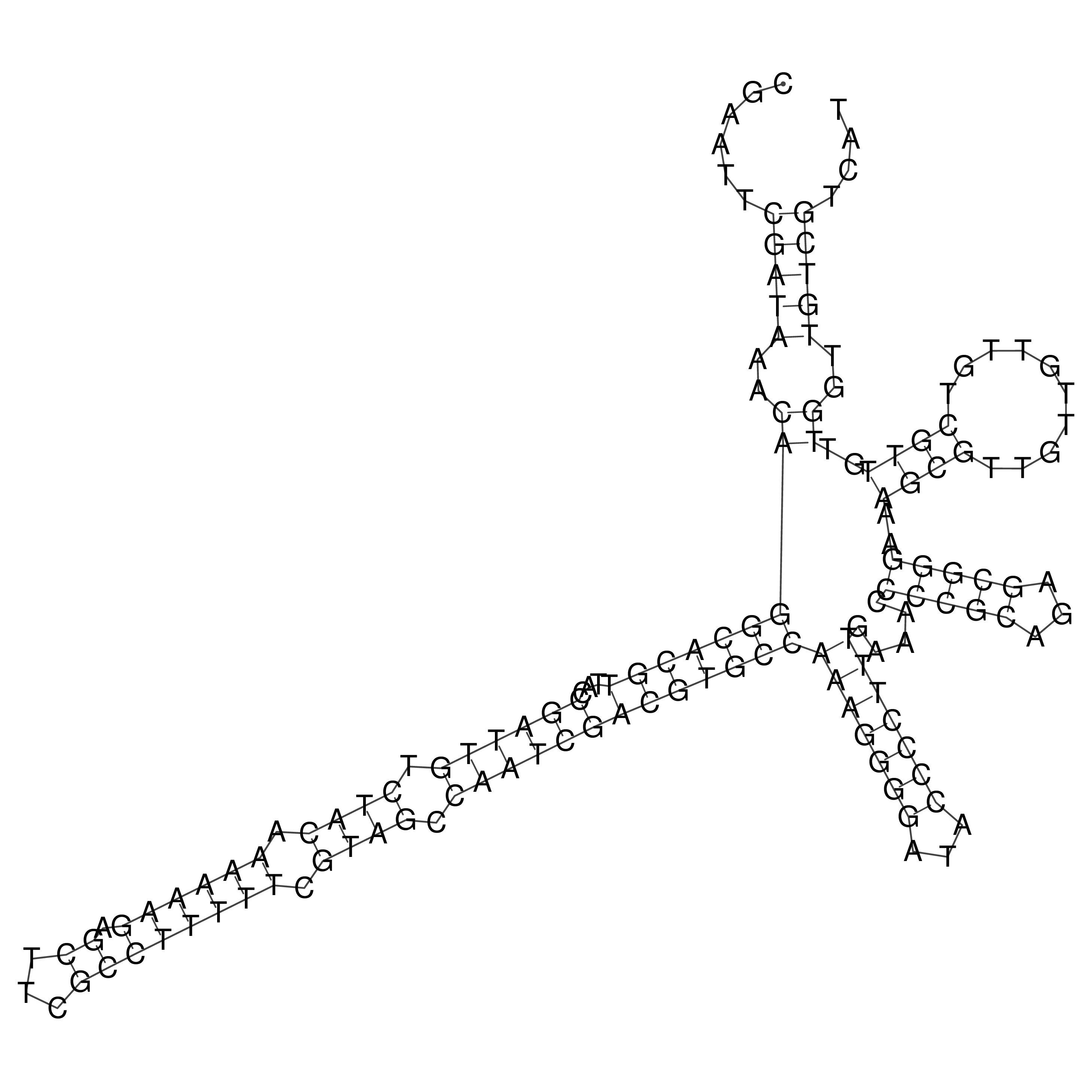
Relaxase
| ID | 566 | GenBank | YP_006963256 |
| Name | Relaxase_pB76-81 |
UniProt ID | I3W0C8 |
| Length | 1249 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1249 a.a. Molecular weight: 138397.68 Da Isoelectric Point: 8.9862
>YP_006963256.1 relaxase, putative (plasmid) [Pseudomonas syringae]
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDAFIVRRELKKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERENNFYRAHGDDYESVHTWQRQRDNQRSPA
ETAGRRTAAPAHGLQDLSVSDVADHRQAGPTRSRDGALLLPTDAHVHVGQSQPGGDSGLRSSVPGGGRGR
RVGSTAERGRGGSQPAAVSQETKGTATPAGATGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSHPQVADQLPQATAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKALVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHKPQPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPGREGVRDQVKRIMDHFAKQVDPAAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADRLGATVPIEALYGQGKTADQVSQALTAQL
DTVPESERIAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAANSAASATSEAQANVPTPSDAVPEPV
SPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMGLRGEDHAIWIIATDDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPQSVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEDAMSDADLKQGDQVRLQDLGTQPVVV
QVIEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAMAAGTPDLSSPDEDDGMSVD
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDAFIVRRELKKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERENNFYRAHGDDYESVHTWQRQRDNQRSPA
ETAGRRTAAPAHGLQDLSVSDVADHRQAGPTRSRDGALLLPTDAHVHVGQSQPGGDSGLRSSVPGGGRGR
RVGSTAERGRGGSQPAAVSQETKGTATPAGATGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSHPQVADQLPQATAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKALVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHKPQPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPGREGVRDQVKRIMDHFAKQVDPAAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADRLGATVPIEALYGQGKTADQVSQALTAQL
DTVPESERIAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAANSAASATSEAQANVPTPSDAVPEPV
SPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMGLRGEDHAIWIIATDDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPQSVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEDAMSDADLKQGDQVRLQDLGTQPVVV
QVIEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAMAAGTPDLSSPDEDDGMSVD
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | I3W0C8 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 57686..68029
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTA54_RS00300 (D708_p077) | 53200..53448 | + | 249 | WP_005618754 | type II toxin-antitoxin system prevent-host-death family antitoxin | - |
| HTA54_RS00305 (D708_p078) | 53461..53811 | + | 351 | WP_015061851 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HTA54_RS00310 (D708_p079) | 53914..54687 | - | 774 | WP_172685659 | helix-turn-helix transcriptional regulator | - |
| HTA54_RS00315 (D708_p080) | 54796..55896 | + | 1101 | WP_015061853 | polyamine ABC transporter substrate-binding protein | - |
| HTA54_RS00320 (D708_p081) | 55993..56208 | + | 216 | WP_015061854 | hypothetical protein | - |
| HTA54_RS00325 | 56218..56544 | - | 327 | WP_080231957 | helix-turn-helix transcriptional regulator | - |
| HTA54_RS00330 (D708_p082) | 56541..56717 | - | 177 | WP_172685652 | hypothetical protein | - |
| HTA54_RS00335 (D708_p083) | 56809..57342 | + | 534 | WP_015061856 | transcription termination/antitermination NusG family protein | - |
| HTA54_RS00340 (D708_p084) | 57431..57673 | + | 243 | WP_015061857 | hypothetical protein | - |
| HTA54_RS00345 (D708_p085) | 57686..58387 | + | 702 | WP_015061858 | lytic transglycosylase domain-containing protein | virB1 |
| HTA54_RS00350 (D708_p086) | 58387..58722 | + | 336 | WP_015061859 | hypothetical protein | - |
| HTA54_RS00355 (D708_p087) | 58735..59223 | + | 489 | WP_015061860 | VirB3 family type IV secretion system protein | virB3 |
| HTA54_RS00360 (D708_p088) | 59150..61624 | + | 2475 | WP_258404827 | conjugal transfer protein | virb4 |
| HTA54_RS00365 (D708_p089) | 61621..62307 | + | 687 | WP_015061862 | P-type DNA transfer protein VirB5 | - |
| HTA54_RS00370 (D708_p090) | 62340..62693 | + | 354 | WP_015061863 | hypothetical protein | - |
| HTA54_RS00375 (D708_p091) | 62704..63636 | + | 933 | WP_015061864 | type IV secretion system protein | virB6 |
| HTA54_RS00380 (D708_p093) | 64044..64832 | + | 789 | WP_015061866 | type IV secretion system protein | virB8 |
| HTA54_RS00385 (D708_p094) | 64822..65631 | + | 810 | WP_015061867 | P-type conjugative transfer protein VirB9 | virB9 |
| HTA54_RS00390 (D708_p095) | 65618..66988 | + | 1371 | WP_015061868 | TrbI/VirB10 family protein | virB10 |
| HTA54_RS00395 (D708_p096) | 66998..68029 | + | 1032 | WP_015061869 | P-type DNA transfer ATPase VirB11 | virB11 |
| HTA54_RS00400 (D708_p097) | 68039..68395 | + | 357 | WP_015061870 | hypothetical protein | - |
| HTA54_RS00405 (D708_p098) | 68515..68832 | + | 318 | WP_015061871 | hypothetical protein | - |
| HTA54_RS00410 (D708_p099) | 68842..69075 | + | 234 | WP_015061872 | hypothetical protein | - |
| HTA54_RS00415 (D708_p100) | 69075..70727 | + | 1653 | WP_015061873 | type IV secretory system conjugative DNA transfer family protein | - |
| HTA54_RS00420 (D708_p101) | 70769..71131 | + | 363 | WP_015061874 | TrbM/KikA/MpfK family conjugal transfer protein | - |
Host bacterium
| ID | 956 | GenBank | NC_019328 |
| Plasmid name | pB76-81 | Incompatibility group | _ |
| Plasmid size | 76232 bp | Coordinate of oriT [Strand] | 783..928 [+] |
| Host baterium | Pseudomonas syringae |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |