Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100474 |
| Name | oriT_pSH146_87 |
| Organism | Salmonella enterica subsp. enterica serovar Heidelberg |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_019131 (32744..32853 [-], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pSH146_87
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file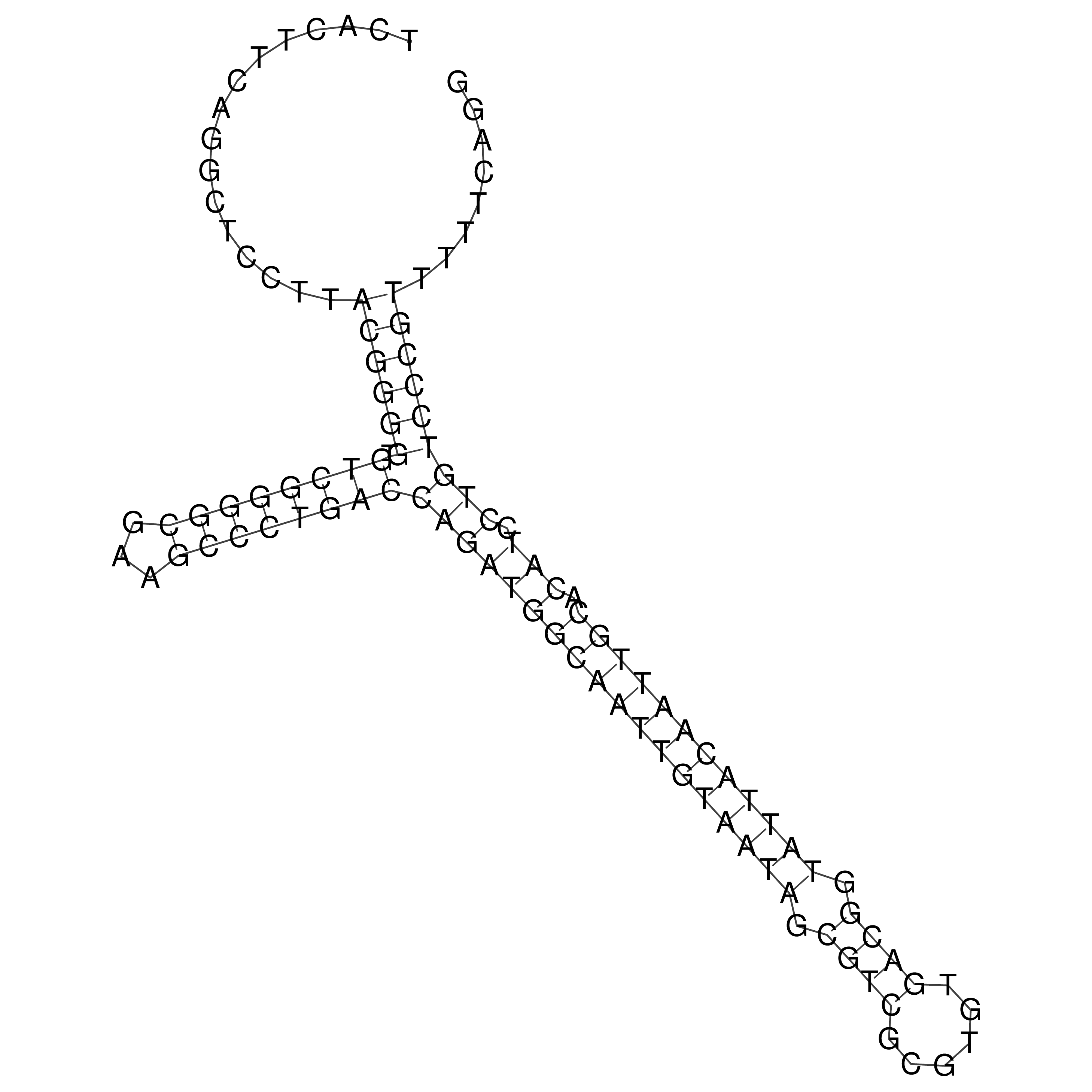
Relaxase
| ID | 545 | GenBank | YP_006957829 |
| Name | Relaxase_pSH146_87 |
UniProt ID | K0H4W4 |
| Length | 871 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 871 a.a. Molecular weight: 100800.84 Da Isoelectric Point: 7.2031
>YP_006957829.1 relaxase (plasmid) [Salmonella enterica subsp. enterica serovar Heidelberg]
MTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVDVMKDGCEWVNFYGVTCFHNCTSLETAAA
DMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVRHTLKSLGLADHQYVSAVHTDTDNLHVHV
AVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCWVHAPGNRIVRKTAVERDRQNAWTRGKKQ
TFREYIAQTAVAGLRSEPVHDWLSMHRRLAEDGLYLSQMDGKFLVMDGWDRNREGVQLDSFGPSWCAEKL
MKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETESLQQYACRHLGERLPEMAREGRLENCQA
IHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLTLDNVNQLDGGWQPVPTDIFLQVTPTERF
RGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGYSISHCSPQIEEMITQGEFTWQRCHELFA
QQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEPQAGPFVSAPADLFDRVQPESRYNPELAV
SDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRKPDLRYGERCREIHQACRLRKSHIRAQYD
DPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYPPSYRQWVEIQAAQGDRAAVSQLRGWDYR
DRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGSVRFRDRRTGEFVCTDYGDRVVFRNHHDR
NALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAWHNVTGRTGHEDYRITRPDVDHHREGSER
YYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVDVMKDGCEWVNFYGVTCFHNCTSLETAAA
DMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVRHTLKSLGLADHQYVSAVHTDTDNLHVHV
AVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCWVHAPGNRIVRKTAVERDRQNAWTRGKKQ
TFREYIAQTAVAGLRSEPVHDWLSMHRRLAEDGLYLSQMDGKFLVMDGWDRNREGVQLDSFGPSWCAEKL
MKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETESLQQYACRHLGERLPEMAREGRLENCQA
IHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLTLDNVNQLDGGWQPVPTDIFLQVTPTERF
RGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGYSISHCSPQIEEMITQGEFTWQRCHELFA
QQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEPQAGPFVSAPADLFDRVQPESRYNPELAV
SDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRKPDLRYGERCREIHQACRLRKSHIRAQYD
DPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYPPSYRQWVEIQAAQGDRAAVSQLRGWDYR
DRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGSVRFRDRRTGEFVCTDYGDRVVFRNHHDR
NALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAWHNVTGRTGHEDYRITRPDVDHHREGSER
YYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K0H4W4 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 38213..76514
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTB66_RS00225 (pSH146_87_62) | 35929..38220 | - | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
| HTB66_RS00230 (pSH146_87_63) | 38213..39283 | - | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| HTB66_RS00235 (pSH146_87_64) | 39302..40510 | - | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| HTB66_RS00240 (pSH146_87_65) | 40802..40954 | + | 153 | WP_001303307 | Hok/Gef family protein | - |
| HTB66_RS00245 (pSH146_87_66) | 41026..41277 | - | 252 | WP_001291964 | hypothetical protein | - |
| HTB66_RS00510 | 41776..41871 | + | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| HTB66_RS00250 | 41936..42220 | - | 285 | WP_172684476 | hypothetical protein | - |
| HTB66_RS00255 (pSH146_87_70) | 42504..42713 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| HTB66_RS00260 (pSH146_87_71) | 42785..43459 | - | 675 | WP_223866377 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HTB66_RS00265 (pSH146_87_72) | 43509..45677 | - | 2169 | WP_000698357 | DotA/TraY family protein | traY |
| HTB66_RS00270 (pSH146_87_73) | 45774..46358 | - | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| HTB66_RS00275 (pSH146_87_74) | 46387..47589 | - | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| HTB66_RS00280 (pSH146_87_75) | 47556..48170 | - | 615 | WP_000337399 | IncI1-type conjugal transfer protein TraV | traV |
| HTB66_RS00285 (pSH146_87_77) | 48170..51214 | - | 3045 | WP_001024782 | IncI1-type conjugal transfer protein TraU | traU |
| HTB66_RS00290 (pSH146_87_78) | 51304..52104 | - | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| HTB66_RS00295 (pSH146_87_79) | 52088..52276 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| HTB66_RS00300 (pSH146_87_80) | 52340..52744 | - | 405 | WP_000086959 | IncI1-type conjugal transfer protein TraR | traR |
| HTB66_RS00305 (pSH146_87_81) | 52795..53322 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| HTB66_RS00310 (pSH146_87_82) | 53322..54026 | - | 705 | WP_000801919 | IncI1-type conjugal transfer protein TraP | traP |
| HTB66_RS00315 (pSH146_87_83) | 54026..55315 | - | 1290 | WP_001272000 | conjugal transfer protein TraO | traO |
| HTB66_RS00320 (pSH146_87_84) | 55318..56301 | - | 984 | WP_001191879 | IncI1-type conjugal transfer protein TraN | traN |
| HTB66_RS00325 (pSH146_87_85) | 56312..57004 | - | 693 | WP_000138551 | DotI/IcmL family type IV secretion protein | traM |
| HTB66_RS00330 (pSH146_87_86) | 57001..57348 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| HTB66_RS00335 (pSH146_87_87) | 57366..61133 | - | 3768 | WP_001141541 | LPD7 domain-containing protein | - |
| HTB66_RS00340 (pSH146_87_88) | 61223..61774 | - | 552 | WP_000014584 | phospholipase D family protein | - |
| HTB66_RS00345 (pSH146_87_89) | 61789..62079 | - | 291 | WP_001372180 | hypothetical protein | traK |
| HTB66_RS00350 (pSH146_87_90) | 62076..63224 | - | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| HTB66_RS00355 (pSH146_87_91) | 63221..64039 | - | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| HTB66_RS00360 (pSH146_87_92) | 64036..64494 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| HTB66_RS00365 (pSH146_87_94) | 64889..65473 | - | 585 | WP_000977522 | histidine phosphatase family protein | - |
| HTB66_RS00370 (pSH146_87_95) | 65533..66735 | - | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| HTB66_RS00375 (pSH146_87_96) | 66821..67645 | - | 825 | WP_001238929 | conjugal transfer protein TraE | traE |
| HTB66_RS00380 (pSH146_87_97) | 67796..68950 | - | 1155 | WP_001139958 | site-specific integrase | - |
| HTB66_RS00500 (pSH146_87_98) | 68934..69302 | + | 369 | WP_015059923 | hypothetical protein | - |
| HTB66_RS00395 | 69592..69747 | - | 156 | WP_001302705 | hypothetical protein | - |
| HTB66_RS00515 | 70366..70587 | + | 222 | Protein_79 | prepilin | - |
| HTB66_RS00405 (pSH146_87_101) | 70584..72008 | - | 1425 | WP_001389385 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HTB66_RS00410 (pSH146_87_102) | 72008..72664 | - | 657 | WP_001193553 | prepilin peptidase | - |
| HTB66_RS00415 | 72649..73209 | - | 561 | WP_000014116 | lytic transglycosylase domain-containing protein | virB1 |
| HTB66_RS00420 (pSH146_87_104) | 73219..73833 | - | 615 | WP_000959786 | type 4 pilus major pilin | - |
| HTB66_RS00425 (pSH146_87_105) | 73851..74948 | - | 1098 | WP_001208805 | type II secretion system F family protein | - |
| HTB66_RS00430 (pSH146_87_106) | 74961..76514 | - | 1554 | WP_000362202 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HTB66_RS00435 (pSH146_87_107) | 76525..76977 | - | 453 | WP_001247334 | type IV pilus biogenesis protein PilP | - |
| HTB66_RS00440 (pSH146_87_108) | 76964..78259 | - | 1296 | WP_015059926 | type 4b pilus protein PilO2 | - |
| HTB66_RS00445 (pSH146_87_109) | 78252..79934 | - | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| HTB66_RS00450 (pSH146_87_110) | 79948..80385 | - | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| HTB66_RS00455 (pSH146_87_111) | 80385..81452 | - | 1068 | WP_001302629 | type IV pilus biogenesis lipoprotein PilL | - |
Host bacterium
| ID | 935 | GenBank | NC_019131 |
| Plasmid name | pSH146_87 | Incompatibility group | IncI1 |
| Plasmid size | 86586 bp | Coordinate of oriT [Strand] | 32744..32853 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Heidelberg |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |