Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100458 |
| Name | oriT_pHHA45 |
| Organism | Escherichia coli |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_019098 (24741..25389 [+], 649 nt) |
| oriT length | 649 nt |
| IRs (inverted repeats) | IR1: 198..207, 211..220 (ATTAGGTGTT..AATACCTAAT) IR2:319..324, 330..335 (AAAAAA..TTTTTT) IR3: 393..402, 405..414 (GTATTCATGC..GCATGAATAC) |
| Location of nic site | 301..302 |
| Conserved sequence flanking the nic site |
GGTGT|ATAGC |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 649 nt
>oriT_pHHA45
AGCGCCGCAGATAATCTGACCGATTACCTCCTGAAACCAGGTCTATATAGGCCAAAAGTTCATCTGATACTTTTGCGGTTATTATTGGCATTCAGTCCTCACATTGTGCATTTCTTAAACAAAAGATTGGGATCTAACAAGCTGAAATCTTAGTATTACCAAAGTAATAAAGCAAACTCATTATAAAACAATGAGTTATTAGGTGTTTTTAATACCTAATTATTACCGAATATTGACGCTATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAGGAGCGATCTACGTAGGTTAAGGACTAACTGACTAAAAAGCGTTCAATATTCCGTATTCATGCTTGCATGAATACCAGTACAACACTATTACAACAAAAGTACATCAAAATTACATCAAAAGTACATCACTTGAAGGTTGACAGTACAACAGAATTACATCATTATCTGGTCATGAGGTAGCCAGTACAACAAAAGTACATCAAAAATACATCATAAATACATCAGAAATACATCAAAATTACATCATTCTAAATGAGGGTACTATGAAGCCCAAAAGTATCAGGGCGGCACTTCAGTTGATGTTGCCGG
AGCGCCGCAGATAATCTGACCGATTACCTCCTGAAACCAGGTCTATATAGGCCAAAAGTTCATCTGATACTTTTGCGGTTATTATTGGCATTCAGTCCTCACATTGTGCATTTCTTAAACAAAAGATTGGGATCTAACAAGCTGAAATCTTAGTATTACCAAAGTAATAAAGCAAACTCATTATAAAACAATGAGTTATTAGGTGTTTTTAATACCTAATTATTACCGAATATTGACGCTATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAGGAGCGATCTACGTAGGTTAAGGACTAACTGACTAAAAAGCGTTCAATATTCCGTATTCATGCTTGCATGAATACCAGTACAACACTATTACAACAAAAGTACATCAAAATTACATCAAAAGTACATCACTTGAAGGTTGACAGTACAACAGAATTACATCATTATCTGGTCATGAGGTAGCCAGTACAACAAAAGTACATCAAAAATACATCATAAATACATCAGAAATACATCAAAATTACATCATTCTAAATGAGGGTACTATGAAGCCCAAAAGTATCAGGGCGGCACTTCAGTTGATGTTGCCGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file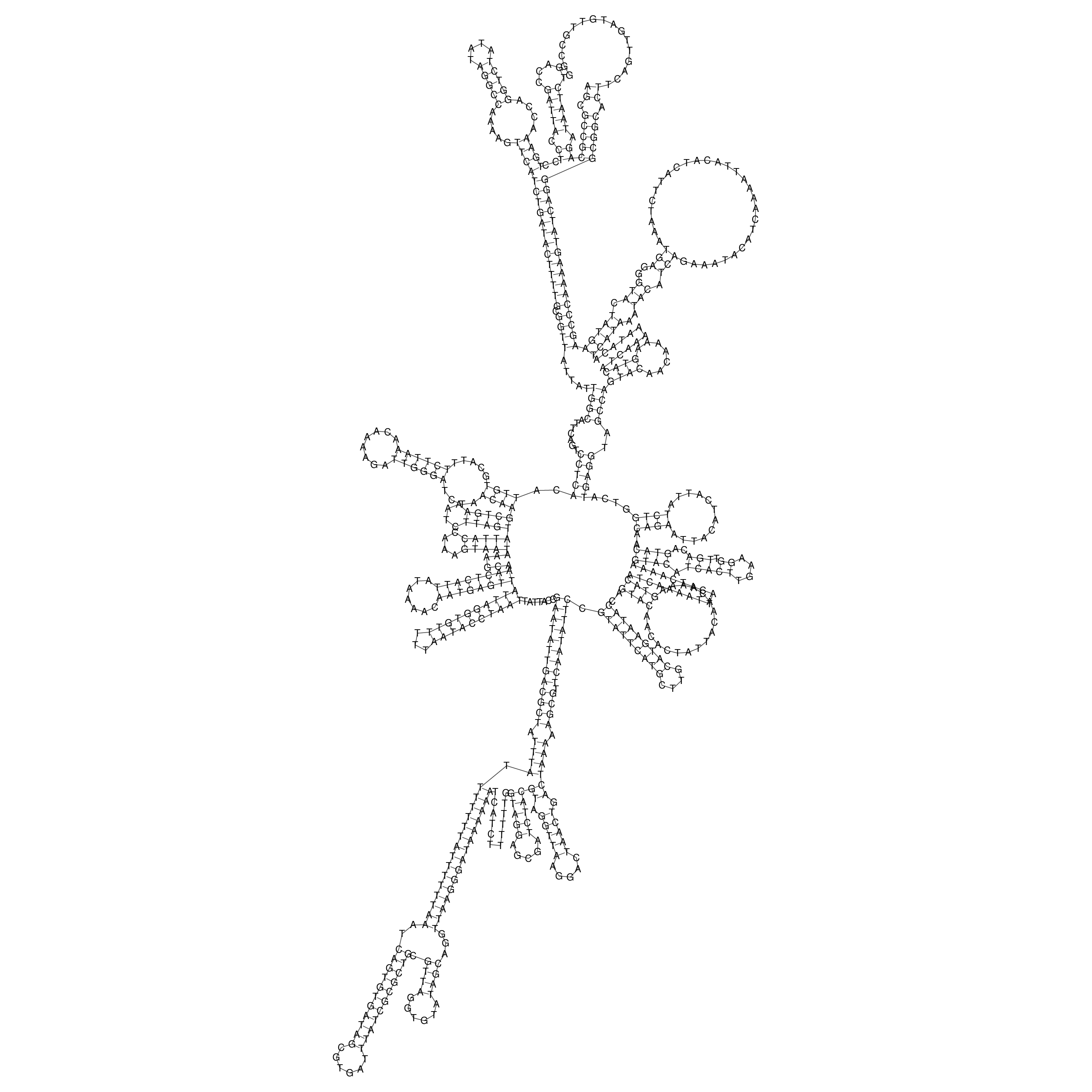
Relaxase
| ID | 529 | GenBank | YP_006954500 |
| Name | TraI_pHHA45 |
UniProt ID | I7EM40 |
| Length | 1078 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120126.36 Da Isoelectric Point: 6.5223
>YP_006954500.1 TraI (plasmid) [Escherichia coli]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLSGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAACALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLSGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAACALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | I7EM40 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 7990..24413
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS959_RS00025 (D616_p79005) | 3264..3614 | + | 351 | Protein_4 | WbuC family cupin fold metalloprotein | - |
| HS959_RS00030 (D616_p79006) | 3661..4536 | - | 876 | WP_013188473 | extended-spectrum class A beta-lactamase CTX-M-1 | - |
| HS959_RS00035 (D616_p79007) | 4894..5598 | + | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| HS959_RS00040 (D616_p79008) | 5660..5857 | - | 198 | WP_000844102 | hypothetical protein | - |
| HS959_RS00045 (D616_p79009) | 5862..6503 | - | 642 | WP_001694850 | restriction endonuclease | - |
| HS959_RS00050 | 6559..6792 | - | 234 | WP_001191790 | hypothetical protein | - |
| HS959_RS00055 (D616_p79010) | 6906..7220 | - | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HS959_RS00060 (D616_p79011) | 7183..7560 | - | 378 | WP_000504251 | hypothetical protein | - |
| HS959_RS00065 (D616_p79012) | 7576..7947 | - | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| HS959_RS00070 (D616_p79013) | 7990..8724 | + | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| HS959_RS00075 (D616_p79014) | 8733..9014 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| HS959_RS00080 (D616_p79015) | 9024..9317 | + | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| HS959_RS00085 (D616_p79016) | 9367..9684 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| HS959_RS00090 (D616_p79017) | 9684..12284 | + | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HS959_RS00095 (D616_p79018) | 12302..13015 | + | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| HS959_RS00100 (D616_p79019) | 13023..13250 | + | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| HS959_RS00105 (D616_p79020) | 13266..14306 | + | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| HS959_RS00110 | 14377..14535 | + | 159 | WP_012561180 | hypothetical protein | - |
| HS959_RS00115 (D616_p79021) | 14525..15223 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| HS959_RS00120 (D616_p79022) | 15234..16118 | + | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| HS959_RS00125 (D616_p79023) | 16118..17278 | + | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| HS959_RS00130 (D616_p79024) | 17320..18315 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HS959_RS00135 (D616_p79025) | 18315..18848 | + | 534 | WP_000731968 | phospholipase D family protein | - |
| HS959_RS00140 (D616_p79026) | 19022..19648 | - | 627 | WP_000434930 | DUF6710 family protein | - |
| HS959_RS00145 (D616_p79027) | 19648..22884 | - | 3237 | WP_015059325 | MobF family relaxase | - |
| HS959_RS00150 (D616_p79028) | 22884..24413 | - | 1530 | WP_000342688 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| HS959_RS00155 (D616_p79029) | 24415..24831 | - | 417 | WP_001532083 | hypothetical protein | - |
| HS959_RS00250 | 25344..25763 | + | 420 | WP_000802909 | plasmid stabilization protein StbA | - |
| HS959_RS00165 (D616_p79031) | 25772..26488 | + | 717 | WP_000861577 | StbB family protein | - |
| HS959_RS00170 (D616_p79032) | 26490..26858 | + | 369 | WP_000414913 | plasmid stabilization protein StbC | - |
| HS959_RS00175 (D616_p79033) | 27040..27384 | + | 345 | WP_015059326 | hypothetical protein | - |
| HS959_RS00180 (D616_p79034) | 27495..27815 | - | 321 | WP_000211833 | protein CcgAII | - |
| HS959_RS00185 (D616_p79035) | 27870..28037 | - | 168 | WP_015059330 | hypothetical protein | - |
| HS959_RS00190 (D616_p79036) | 28869..29378 | - | 510 | WP_000129958 | antirestriction protein ArdA | - |
Host bacterium
| ID | 919 | GenBank | NC_019098 |
| Plasmid name | pHHA45 | Incompatibility group | IncN |
| Plasmid size | 39510 bp | Coordinate of oriT [Strand] | 24741..25389 [+] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | mph(A), blaCTX-M-1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |