Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100392 |
| Name | oriT_pEKO1101 |
| Organism | Escherichia coli KO11FL |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_016904 (31870..31979 [-], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pEKO1101
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file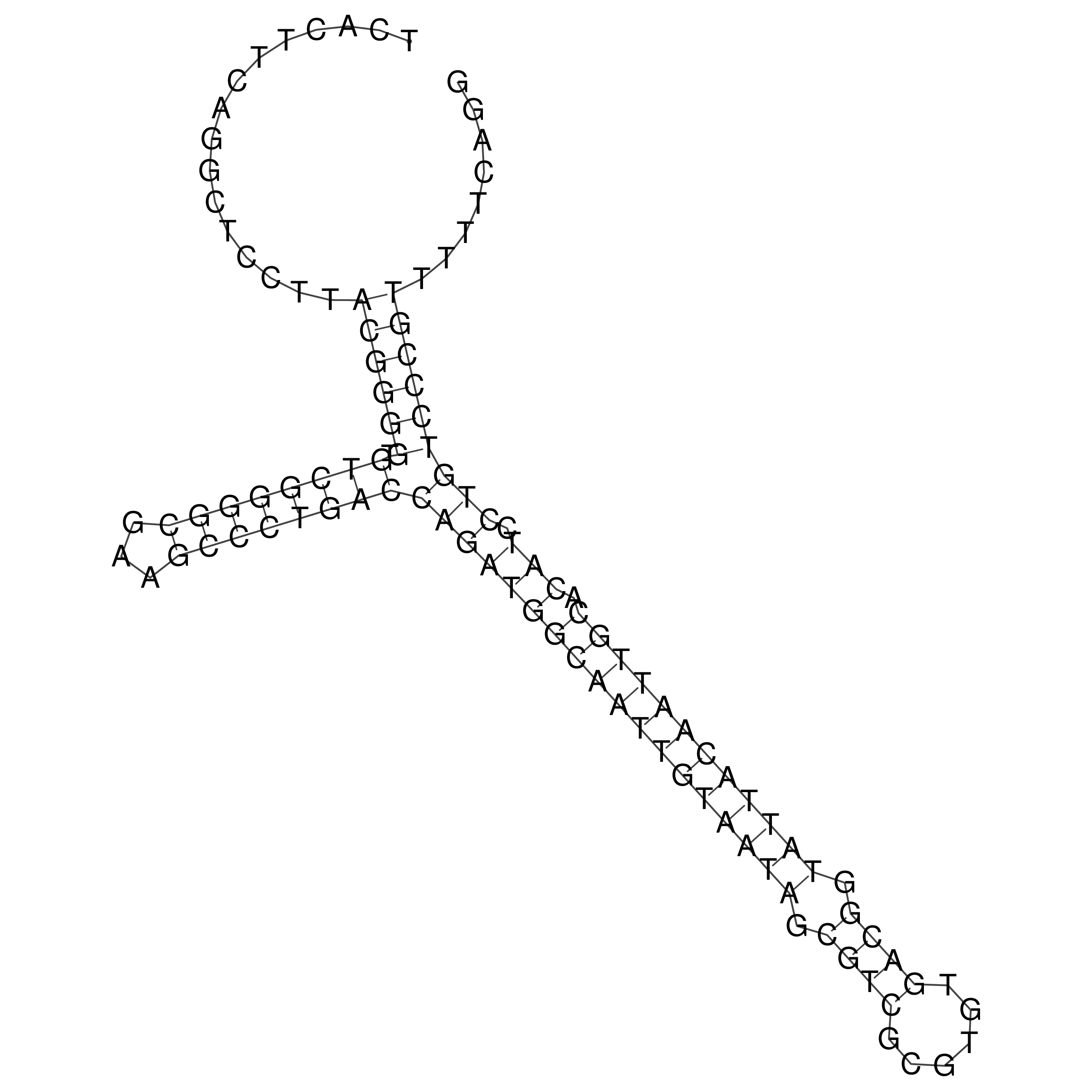
Relaxase
| ID | 463 | GenBank | WP_001350015 |
| Name | NikB_pEKO1101 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104034.43 Da Isoelectric Point: 7.3526
>WP_001350015.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 37339..74503
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EKO11_RS24795 (EKO11_4695) | 35055..37346 | - | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
| EKO11_RS24800 (EKO11_4696) | 37339..38409 | - | 1071 | WP_000151587 | IncI1-type conjugal transfer protein TrbB | trbB |
| EKO11_RS24805 (EKO11_4697) | 38428..39636 | - | 1209 | WP_000121272 | IncI1-type conjugal transfer protein TrbA | trbA |
| EKO11_RS24810 (EKO11_4698) | 39898..40806 | - | 909 | WP_000055084 | hypothetical protein | - |
| EKO11_RS27995 (EKO11_4700) | 41598..41774 | - | 177 | WP_001054904 | hypothetical protein | - |
| EKO11_RS24830 (EKO11_4701) | 41984..42175 | - | 192 | WP_001147208 | hemolysin expression modulator Hha | - |
| EKO11_RS24835 (EKO11_4702) | 42241..42855 | - | 615 | WP_000578649 | plasmid IncI1-type surface exclusion protein ExcA | - |
| EKO11_RS24840 (EKO11_4703) | 42931..45099 | - | 2169 | WP_000698360 | IncI1-type conjugal transfer membrane protein TraY | traY |
| EKO11_RS24845 (EKO11_4704) | 45196..45780 | - | 585 | WP_001037990 | IncI1-type conjugal transfer protein TraX | - |
| EKO11_RS24850 (EKO11_4705) | 45809..47011 | - | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| EKO11_RS24855 (EKO11_4706) | 46978..47592 | - | 615 | WP_000337400 | IncI1-type conjugal transfer protein TraV | traV |
| EKO11_RS24860 (EKO11_4707) | 47592..50636 | - | 3045 | WP_001024787 | IncI1-type conjugal transfer protein TraU | traU |
| EKO11_RS24865 (EKO11_4708) | 50726..51526 | - | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| EKO11_RS24870 (EKO11_4709) | 51510..51698 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| EKO11_RS24875 (EKO11_4710) | 51762..52166 | - | 405 | WP_000086960 | IncI1-type conjugal transfer protein TraR | traR |
| EKO11_RS24880 (EKO11_4711) | 52217..52744 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| EKO11_RS24885 (EKO11_4712) | 52744..53448 | - | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| EKO11_RS24890 (EKO11_4713) | 53448..54737 | - | 1290 | WP_001271991 | conjugal transfer protein TraO | traO |
| EKO11_RS24895 (EKO11_4714) | 54740..55723 | - | 984 | WP_001191878 | IncI1-type conjugal transfer protein TraN | traN |
| EKO11_RS24900 (EKO11_4715) | 55734..56426 | - | 693 | WP_000138549 | DotI/IcmL family type IV secretion protein | traM |
| EKO11_RS24905 (EKO11_4716) | 56423..56770 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| EKO11_RS24910 (EKO11_4717) | 56788..60555 | - | 3768 | WP_001141538 | LPD7 domain-containing protein | - |
| EKO11_RS24915 (EKO11_4718) | 60645..61196 | - | 552 | WP_000014584 | phospholipase D family protein | - |
| EKO11_RS26850 (EKO11_4719) | 61211..61501 | - | 291 | WP_001299214 | hypothetical protein | traK |
| EKO11_RS24920 | 61498..62040 | - | 543 | Protein_70 | Flp pilus assembly complex ATPase component TadA | - |
| EKO11_RS24935 | 63299..63907 | - | 609 | Protein_72 | Flp pilus assembly complex ATPase component TadA | - |
| EKO11_RS24940 (EKO11_4722) | 63904..64722 | - | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| EKO11_RS24945 (EKO11_4723) | 64719..65177 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| EKO11_RS24950 (EKO11_4724) | 65572..66156 | - | 585 | WP_000977526 | histidine phosphatase family protein | - |
| EKO11_RS24955 (EKO11_4725) | 66216..67418 | - | 1203 | WP_000976350 | conjugal transfer protein TraF | - |
| EKO11_RS24960 (EKO11_4726) | 67504..68328 | - | 825 | WP_001238943 | conjugal transfer protein TraE | traE |
| EKO11_RS27440 (EKO11_4727) | 68536..68661 | + | 126 | Protein_78 | transposase | - |
| EKO11_RS24965 (EKO11_4728) | 68718..70010 | - | 1293 | WP_001350014 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| EKO11_RS24970 (EKO11_4729) | 70010..70666 | - | 657 | WP_001193550 | prepilin peptidase | - |
| EKO11_RS24975 (EKO11_4730) | 70651..71211 | - | 561 | WP_000014005 | lytic transglycosylase domain-containing protein | virB1 |
| EKO11_RS24980 (EKO11_4731) | 71221..71835 | - | 615 | WP_000908226 | type 4 pilus major pilin | - |
| EKO11_RS24985 (EKO11_4732) | 71852..72937 | - | 1086 | WP_001208804 | type II secretion system F family protein | - |
| EKO11_RS24990 (EKO11_4733) | 72950..74503 | - | 1554 | WP_000362206 | ATPase, T2SS/T4P/T4SS family | virB11 |
| EKO11_RS24995 (EKO11_4734) | 74514..74966 | - | 453 | WP_001236383 | type IV pilus biogenesis protein PilP | - |
| EKO11_RS25000 (EKO11_4735) | 74953..76248 | - | 1296 | WP_000752792 | type 4b pilus protein PilO2 | - |
| EKO11_RS25005 (EKO11_4736) | 76241..77923 | - | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| EKO11_RS25010 (EKO11_4737) | 77937..78374 | - | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| EKO11_RS25015 (EKO11_4738) | 78374..79441 | - | 1068 | WP_001350013 | type IV pilus biogenesis lipoprotein PilL | - |
Host bacterium
| ID | 853 | GenBank | NC_016904 |
| Plasmid name | pEKO1101 | Incompatibility group | IncI |
| Plasmid size | 103795 bp | Coordinate of oriT [Strand] | 31870..31979 [-] |
| Host baterium | Escherichia coli KO11FL |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |