Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100356 |
| Name | oriT_pO26-CRL |
| Organism | Escherichia coli O26:H- |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_013728 (68546..68637 [-], 92 nt) |
| oriT length | 92 nt |
| IRs (inverted repeats) | IR1: 7..15, 17..25 (GTCGGGGCT..AGCCCTGAC) IR2: 32..49, 52..69 (GTAATCGTATAGCCGTGC..GCGCGGTTATACGATTAC) |
| Location of nic site | 77..78 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 92 nt
>oriT_pO26-CRL
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTTTCAGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file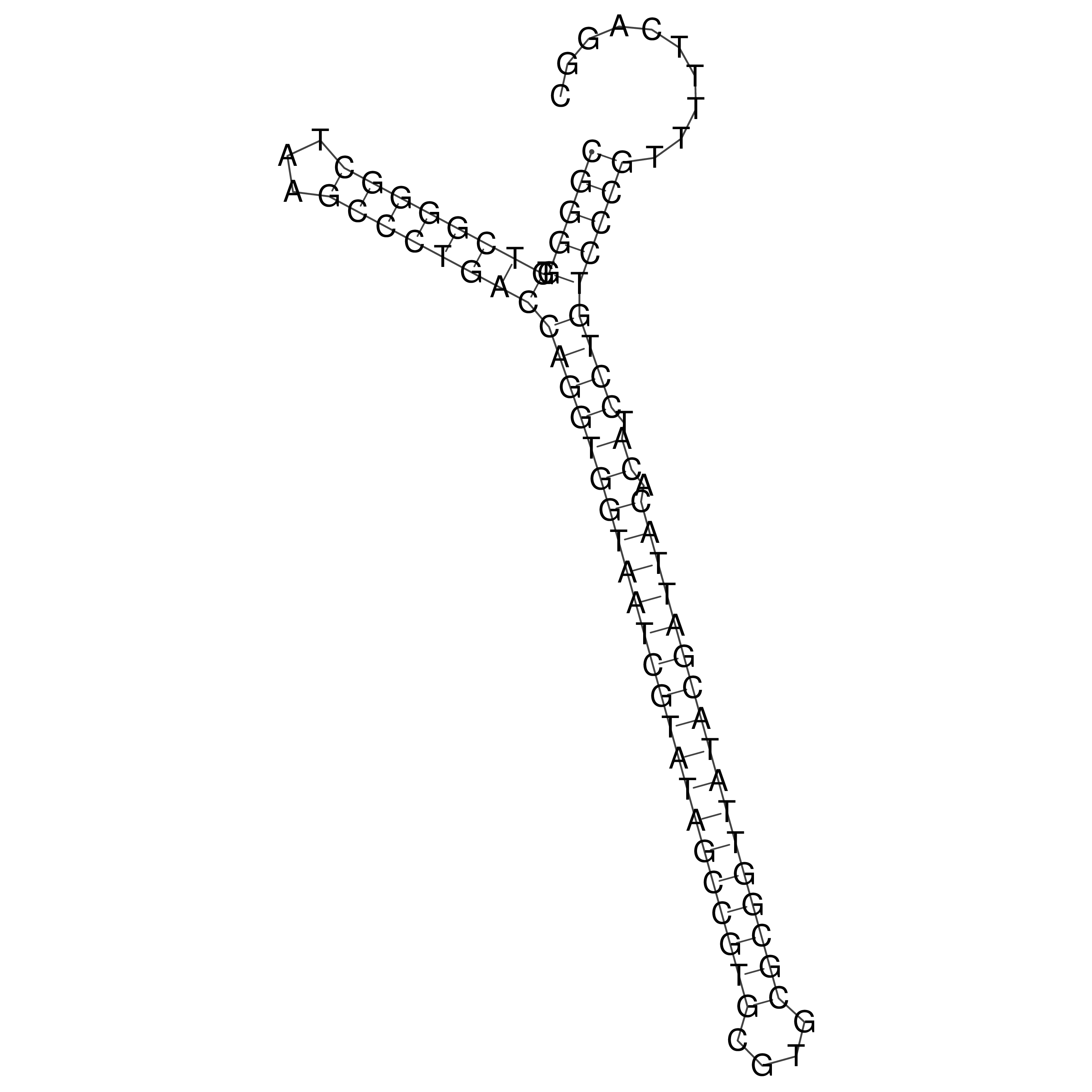
Relaxase
| ID | 427 | GenBank | YP_003377894 |
| Name | Nuclease_pO26-CRL |
UniProt ID | D2WFH4 |
| Length | 906 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 105076.81 Da Isoelectric Point: 8.1396
>YP_003377894.1 relaxase/mobilization nuclease (plasmid) [Escherichia coli O26:H-]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELEIKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIVQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPLWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLATAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSVSHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELEIKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIVQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPLWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPEMARNGELESCLDVHRTLATAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGRNLGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSVSHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D2WFH4 |
Host bacterium
| ID | 817 | GenBank | NC_013728 |
| Plasmid name | pO26-CRL | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 111481 bp | Coordinate of oriT [Strand] | 68546..68637 [-] |
| Host baterium | Escherichia coli O26:H- |
Cargo genes
| Drug resistance gene | dfrA5, blaTEM-1B, sul2, aph(3'')-Ib, aph(6)-Id, aph(3')-Ia |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |