Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100353 |
| Name | oriT_pPR9 |
| Organism | Staphylococcus aureus |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_013653 (20522..20557 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 31..32 |
| Conserved sequence flanking the nic site |
TAAGTGCGCC|CT |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 36 nt
>oriT_pPR9
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file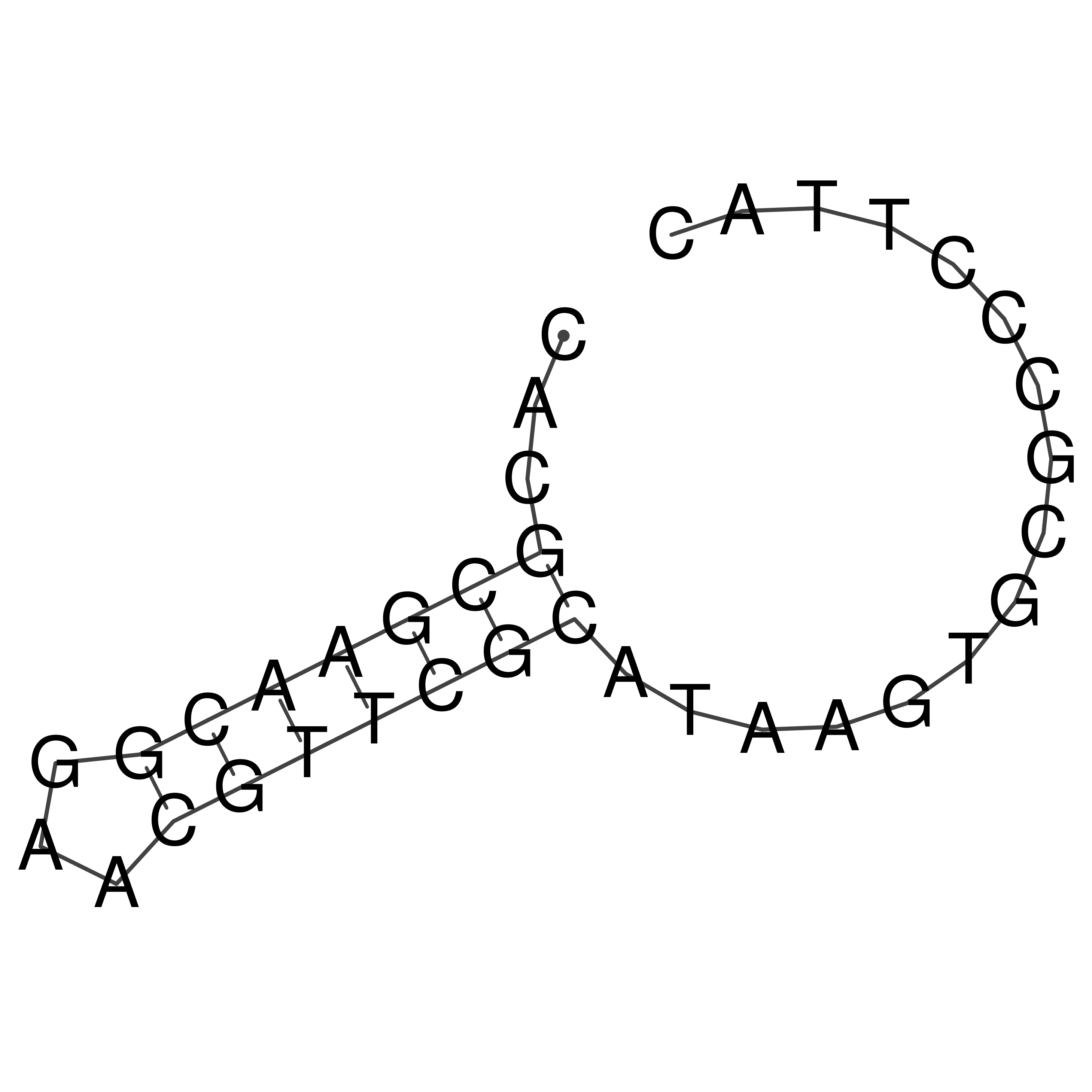
Relaxase
| ID | 424 | GenBank | YP_003347965 |
| Name | Nes_pPR9 |
UniProt ID | D2KNY7 |
| Length | 665 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 78929.92 Da Isoelectric Point: 5.5367
>YP_003347965.1 oriT nickase Nes (plasmid) [Staphylococcus aureus]
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNSYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGQKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYEESERLYDLKQKTALITDDEDRLNKIENIEDRQRKLESINEVFEKQSKIFFDKNYPDQKLNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGSENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVLGAIQKKLLSEY
DFDYSDNNDLKHLYQESYEVGDEISKDNIEEFYEGNDILVDKETYNNYNKKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNSYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGQKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYEESERLYDLKQKTALITDDEDRLNKIENIEDRQRKLESINEVFEKQSKIFFDKNYPDQKLNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGSENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVLGAIQKKLLSEY
DFDYSDNNDLKHLYQESYEVGDEISKDNIEEFYEGNDILVDKETYNNYNKKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D2KNY7 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26396..39480
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS657_RS00105 (HUNSC491_pPR9_p20) | 21537..22559 | + | 1023 | WP_224684886 | parM protein | - |
| HS657_RS00110 (HUNSC491_pPR9_p21) | 22562..22891 | + | 330 | WP_000358311 | recombinase | - |
| HS657_RS00115 (HUNSC491_pPR9_p22) | 22895..23155 | - | 261 | WP_001008214 | helix-turn-helix transcriptional regulator | - |
| HS657_RS00215 (HUNSC491_pPR9_p23) | 23537..24562 | + | 1026 | WP_012896704 | replication initiator protein | - |
| HS657_RS00125 (HUNSC491_pPR9_p24) | 24667..24855 | - | 189 | WP_012263508 | hypothetical protein | - |
| HS657_RS00130 (HUNSC491_pPR9_p25) | 25028..26002 | + | 975 | WP_012896705 | conjugative transfer protein TrsA | - |
| HS657_RS00135 (HUNSC491_pPR9_p26) | 26019..26336 | + | 318 | WP_012896706 | CagC family type IV secretion system protein | - |
| HS657_RS00140 (HUNSC491_pPR9_p27) | 26396..26803 | + | 408 | WP_061739438 | hypothetical protein | trsC |
| HS657_RS00145 (HUNSC491_pPR9_p28) | 26790..27473 | + | 684 | WP_012263511 | TrsD/TraD family conjugative transfer protein | trsD |
| HS657_RS00150 (HUNSC491_pPR9_p29) | 27488..29506 | + | 2019 | WP_012896709 | DUF87 domain-containing protein | virb4 |
| HS657_RS00155 (HUNSC491_pPR9_p30) | 29518..30798 | + | 1281 | WP_012896710 | conjugal transfer protein TraF | trsF |
| HS657_RS00160 (HUNSC491_pPR9_p31) | 30816..31892 | + | 1077 | WP_012896711 | CHAP domain-containing protein | trsG |
| HS657_RS00165 (HUNSC491_pPR9_p32) | 31902..32387 | + | 486 | WP_012896712 | TrsH/TraH family protein | - |
| HS657_RS00170 (HUNSC491_pPR9_p33) | 32403..34505 | + | 2103 | WP_012896713 | DNA topoisomerase III | - |
| HS657_RS00175 (HUNSC491_pPR9_p34) | 34521..34985 | + | 465 | WP_012896714 | hypothetical protein | trsJ |
| HS657_RS00180 (HUNSC491_pPR9_p35) | 34982..36628 | + | 1647 | WP_012896715 | VirD4-like conjugal transfer protein, CD1115 family | - |
| HS657_RS00185 (HUNSC491_pPR9_p36) | 36706..37623 | + | 918 | WP_012896716 | conjugal transfer protein TrbL family protein | trsL |
| HS657_RS00190 (HUNSC491_pPR9_p37) | 37640..38032 | + | 393 | WP_012896717 | single-stranded DNA-binding protein | - |
| HS657_RS00195 (HUNSC491_pPR9_p38) | 38089..38820 | + | 732 | WP_012896718 | hypothetical protein | - |
| HS657_RS00200 (HUNSC491_pPR9_p39) | 38857..39480 | + | 624 | WP_061739436 | hypothetical protein | traP |
| HS657_RS00205 (HUNSC491_pPR9_p40) | 39496..40110 | + | 615 | WP_012896720 | hypothetical protein | - |
| HS657_RS00210 (HUNSC491_pPR9_p41) | 40179..41450 | + | 1272 | WP_012896721 | hypothetical protein | - |
Host bacterium
| ID | 814 | GenBank | NC_013653 |
| Plasmid name | pPR9 | Incompatibility group | _ |
| Plasmid size | 41715 bp | Coordinate of oriT [Strand] | 20522..20557 [+] |
| Host baterium | Staphylococcus aureus |
Cargo genes
| Drug resistance gene | mupA, blaZ |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |