Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100326 |
| Name | oriT_pR132503 |
| Organism | Rhizobium leguminosarum bv. trifolii WSM1325 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_012853 (102640..102680 [-], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | 8..9 |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 41 nt
>oriT_pR132503
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file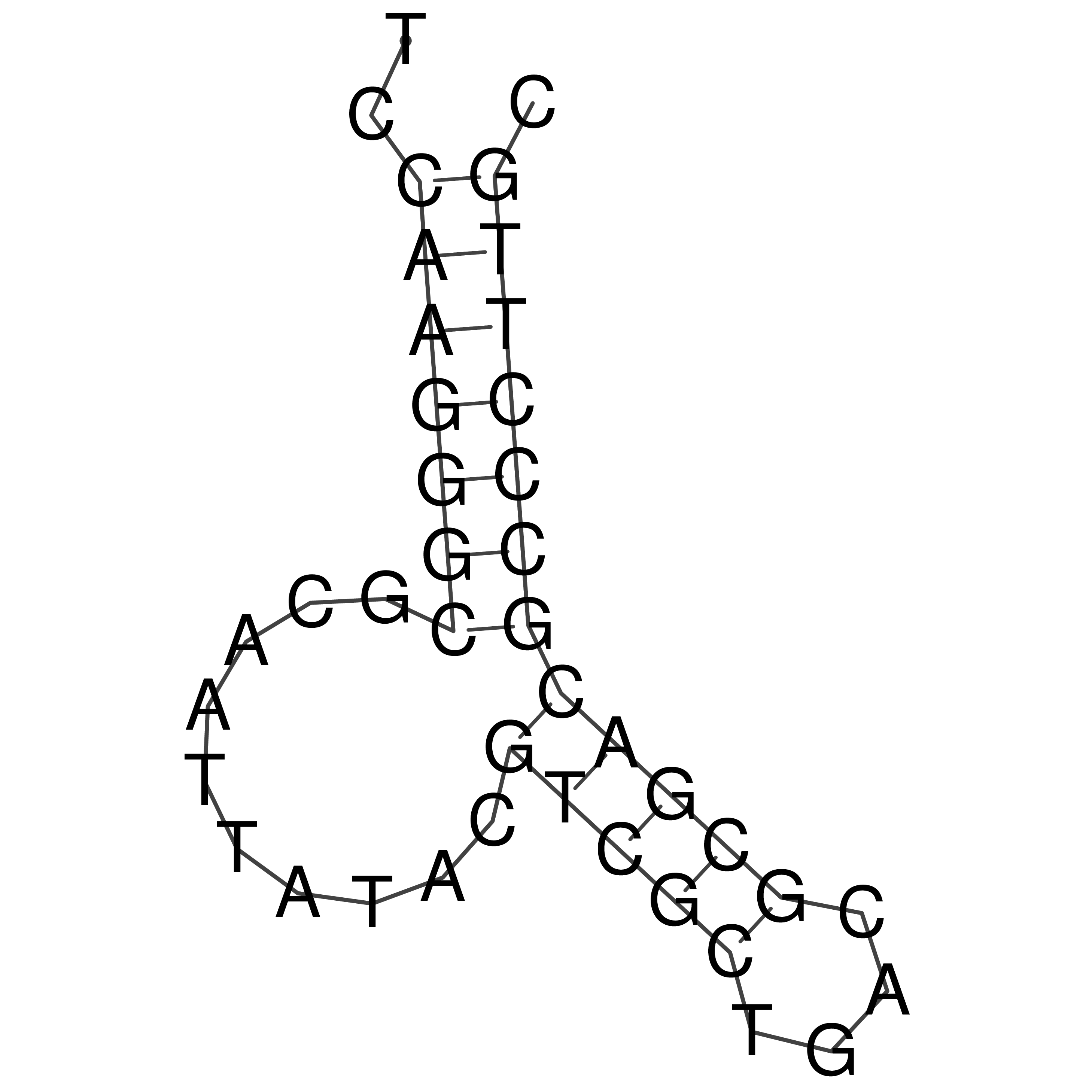
Relaxase
| ID | 397 | GenBank | WP_012760269 |
| Name | TraA_pR132503 |
UniProt ID | C6B8U2 |
| Length | 1107 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 123028.43 Da Isoelectric Point: 10.3023
>WP_012760269.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLYEEFVIPAGAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSAEQSIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTERGFGAKKIAVTSPDGNPMRNDAGKIVYELWAGSADDFNTIRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARSSAWSPKLERIELQEDRRTENARRIQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDAALFQNLMVRILQSPEALRLERDRIDFATGIRTPAKYTTREM
IRLEAEMVSRAIWLSGRASHGVREAVLEATFARHARLSDEQRTAIERVAGGERIAAVIGRAGAGKTTMMK
AAREAWEASGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQSRDQIDTRTVFVLDEAGMVSSRQM
ALFVEAVSKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELKTIYRQRQQWMRDASLDLGRGNVGQA
VDAYRAHGHVRGLDLKAQAVESLIADWDRDYDPSKSSLILAHLRRDVRMLNDMARAKLVERGILADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIDAAAGRLVVALGEGEHRRLVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAIYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KSSLYGQALRFAEARGLHLMNVARTIAHDRLQWVVRQKQKLADLGARLAAIGVILGRAASAKTSPKSKAK
ESKPMVSGISTFPKSIDQSVEDKLAADPGLKKQWEDVSTRFHLVYAQPESAFKAVNVDAMLKDQAIAKTT
LAKIAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEIERKHETEERAVRRQVS
VDIPALSAPAKQTLERVRDAIDRDDLPAGLEYALAVKMVKAELDRFAKAVSQRFGERTFLGIASKDPSGE
TFKSVTAGMNPAQKAEVQAAWNSMRTIQQLSAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLYEEFVIPAGAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSAEQSIALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTERGFGAKKIAVTSPDGNPMRNDAGKIVYELWAGSADDFNTIRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARSSAWSPKLERIELQEDRRTENARRIQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDAALFQNLMVRILQSPEALRLERDRIDFATGIRTPAKYTTREM
IRLEAEMVSRAIWLSGRASHGVREAVLEATFARHARLSDEQRTAIERVAGGERIAAVIGRAGAGKTTMMK
AAREAWEASGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQSRDQIDTRTVFVLDEAGMVSSRQM
ALFVEAVSKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELKTIYRQRQQWMRDASLDLGRGNVGQA
VDAYRAHGHVRGLDLKAQAVESLIADWDRDYDPSKSSLILAHLRRDVRMLNDMARAKLVERGILADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIDAAAGRLVVALGEGEHRRLVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAIYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KSSLYGQALRFAEARGLHLMNVARTIAHDRLQWVVRQKQKLADLGARLAAIGVILGRAASAKTSPKSKAK
ESKPMVSGISTFPKSIDQSVEDKLAADPGLKKQWEDVSTRFHLVYAQPESAFKAVNVDAMLKDQAIAKTT
LAKIAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEIERKHETEERAVRRQVS
VDIPALSAPAKQTLERVRDAIDRDDLPAGLEYALAVKMVKAELDRFAKAVSQRFGERTFLGIASKDPSGE
TFKSVTAGMNPAQKAEVQAAWNSMRTIQQLSAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | C6B8U2 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 68435..77902
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RLEG_RS30290 (Rleg_5478) | 63685..64950 | - | 1266 | WP_041935847 | plasmid replication protein RepC | - |
| RLEG_RS30295 (Rleg_5479) | 65188..66228 | - | 1041 | WP_017997447 | plasmid partitioning protein RepB | - |
| RLEG_RS30300 (Rleg_5480) | 66213..67430 | - | 1218 | WP_012760236 | plasmid partitioning protein RepA | - |
| RLEG_RS30305 (Rleg_5481) | 67795..68421 | + | 627 | WP_012760237 | acyl-homoserine-lactone synthase | - |
| RLEG_RS30310 (Rleg_5482) | 68435..69400 | + | 966 | WP_012760238 | P-type conjugative transfer ATPase TrbB | virB11 |
| RLEG_RS30315 (Rleg_5483) | 69390..69770 | + | 381 | WP_012760239 | TrbC/VirB2 family protein | virB2 |
| RLEG_RS30320 (Rleg_5484) | 69760..70059 | + | 300 | WP_012760240 | conjugal transfer protein TrbD | virB3 |
| RLEG_RS30325 (Rleg_5485) | 70069..72516 | + | 2448 | WP_012760241 | conjugal transfer protein TrbE | virb4 |
| RLEG_RS30330 (Rleg_5486) | 72488..73288 | + | 801 | WP_012760242 | P-type conjugative transfer protein TrbJ | virB5 |
| RLEG_RS30335 (Rleg_5488) | 73508..74656 | + | 1149 | WP_012760244 | P-type conjugative transfer protein TrbL | virB6 |
| RLEG_RS30340 (Rleg_5489) | 74676..75338 | + | 663 | WP_012760245 | conjugal transfer protein TrbF | virB8 |
| RLEG_RS30345 (Rleg_5490) | 75355..76176 | + | 822 | WP_012760246 | P-type conjugative transfer protein TrbG | virB9 |
| RLEG_RS30350 (Rleg_5491) | 76179..76607 | + | 429 | WP_012760247 | conjugal transfer protein TrbH | - |
| RLEG_RS30355 (Rleg_5492) | 76622..77902 | + | 1281 | WP_012760248 | IncP-type conjugal transfer protein TrbI | virB10 |
| RLEG_RS30360 (Rleg_5493) | 77928..78638 | - | 711 | WP_012760249 | autoinducer binding domain-containing protein | - |
| RLEG_RS36615 (Rleg_5494) | 78895..79074 | + | 180 | Protein_72 | IS6 family transposase | - |
| RLEG_RS30365 (Rleg_5495) | 79546..80628 | + | 1083 | WP_012760250 | hypothetical protein | - |
| RLEG_RS30370 (Rleg_5496) | 80625..81536 | + | 912 | WP_012760251 | hypothetical protein | - |
| RLEG_RS36620 | 81735..81941 | + | 207 | WP_210162204 | hypothetical protein | - |
Host bacterium
| ID | 787 | GenBank | NC_012853 |
| Plasmid name | pR132503 | Incompatibility group | _ |
| Plasmid size | 516088 bp | Coordinate of oriT [Strand] | 102640..102680 [-] |
| Host baterium | Rhizobium leguminosarum bv. trifolii WSM1325 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |