Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100322 |
| Name | oriT2_pO26-Vir |
| Organism | Escherichia coli |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_012487 ( 150410..150495 [+], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | IR1: 7..15, 17..25 (GTCGGGGCT..AGCCCTGAC) IR2: 32..49, 52..69 (GTAATCGTATAGCCGTGC..GCGCGGTTATACGATTAC) |
| Location of nic site | 77..78 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 86 nt
>oriT2_pO26-Vir
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file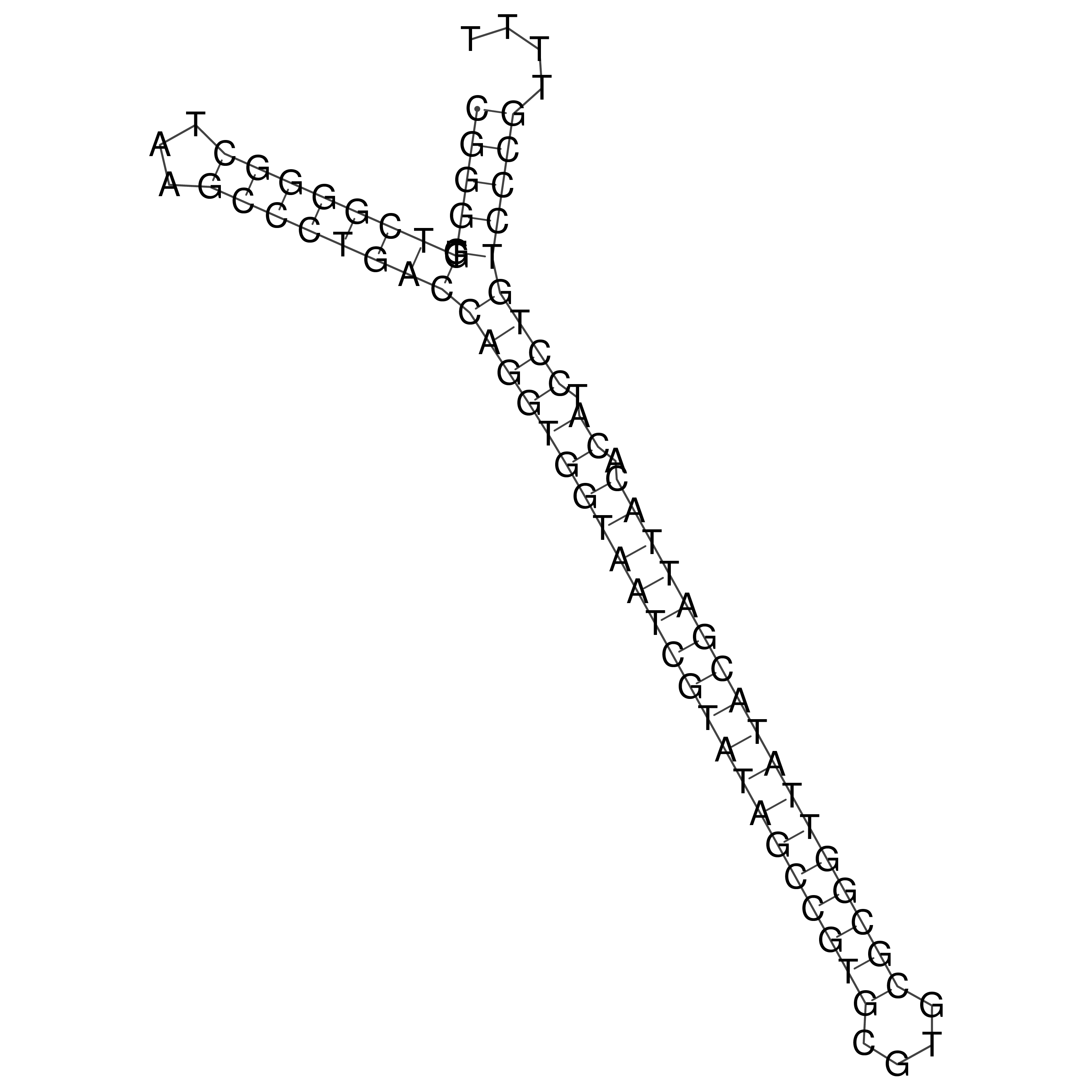
Relaxase
| ID | 393 | GenBank | YP_002756716 |
| Name | Relaxase_pO26-Vir |
UniProt ID | Q68J83 |
| Length | 906 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104908.50 Da Isoelectric Point: 8.2690
>YP_002756716.1 relaxase (plasmid) [Escherichia coli]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVEAPGRYDPEAINVDIRPEKVAETE
SLKQYASRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVEAPGRYDPEAINVDIRPEKVAETE
SLKQYASRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q68J83 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 105548..144829
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKK63_RS00535 (p026VIR_p115) | 101592..102029 | + | 438 | WP_000539951 | type IV pilus biogenesis protein PilM | - |
| HKK63_RS00540 (p026VIR_p116) | 102061..103680 | + | 1620 | WP_001035565 | PilN family type IVB pilus formation outer membrane protein | - |
| HKK63_RS00545 (p026VIR_p117) | 103701..104996 | + | 1296 | WP_000129883 | type 4b pilus protein PilO2 | - |
| HKK63_RS00550 (p026VIR_p118) | 104986..105444 | + | 459 | WP_000095886 | type IV pilus biogenesis protein PilP | - |
| HKK63_RS00555 (p026VIR_p119) | 105548..107056 | + | 1509 | WP_000880806 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HKK63_RS00560 (p026VIR_p120) | 107058..108152 | + | 1095 | WP_000697154 | type II secretion system F family protein | - |
| HKK63_RS00565 (p026VIR_p121) | 108214..108750 | + | 537 | WP_000111521 | type 4 pilus major pilin | - |
| HKK63_RS00570 (p026VIR_p122) | 108795..109280 | + | 486 | WP_001080767 | lytic transglycosylase domain-containing protein | virB1 |
| HKK63_RS00575 (p026VIR_p123) | 109386..109922 | + | 537 | WP_023649457 | prepilin peptidase | - |
| HKK63_RS00580 (p026VIR_p124) | 109940..111301 | + | 1362 | WP_001168171 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HKK63_RS00915 | 111415..111609 | + | 195 | Protein_112 | integrase | - |
| HKK63_RS00585 (p026VIR_p126) | 111774..112595 | + | 822 | WP_000845373 | hypothetical protein | traE |
| HKK63_RS00590 (p026VIR_p127) | 112697..113899 | + | 1203 | WP_000979993 | conjugal transfer protein TraF | - |
| HKK63_RS00595 (p026VIR_p128) | 114003..114461 | + | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| HKK63_RS00600 (p026VIR_p129) | 114458..115294 | + | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| HKK63_RS00605 (p026VIR_p130) | 115278..116426 | + | 1149 | WP_000775210 | plasmid transfer ATPase TraJ | virB11 |
| HKK63_RS00890 | 116423..116713 | + | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| HKK63_RS00610 (p026VIR_p132) | 116777..120838 | + | 4062 | WP_001441856 | LPD7 domain-containing protein | - |
| HKK63_RS00615 (p026VIR_p133) | 120855..121205 | + | 351 | WP_001056367 | hypothetical protein | traL |
| HKK63_RS00620 (p026VIR_p134) | 121217..121912 | + | 696 | WP_001287545 | DotI/IcmL family type IV secretion protein | traM |
| HKK63_RS00625 (p026VIR_p135) | 121923..122897 | + | 975 | WP_000547385 | DotH/IcmK family type IV secretion protein | traN |
| HKK63_RS00630 (p026VIR_p136) | 122901..124238 | + | 1338 | WP_001272018 | conjugal transfer protein TraO | traO |
| HKK63_RS00635 (p026VIR_p137) | 124235..124948 | + | 714 | WP_000787003 | conjugal transfer protein TraP | traP |
| HKK63_RS00640 (p026VIR_p138) | 124945..125475 | + | 531 | WP_000987021 | conjugal transfer protein TraQ | traQ |
| HKK63_RS00645 (p026VIR_p139) | 125522..125920 | + | 399 | WP_000088813 | DUF6750 family protein | traR |
| HKK63_RS00655 (p026VIR_p141) | 126143..126961 | + | 819 | WP_012680981 | hypothetical protein | traT |
| HKK63_RS00660 (p026VIR_p142) | 126954..127154 | - | 201 | WP_179894028 | hypothetical protein | - |
| HKK63_RS00665 (p026VIR_p143) | 127256..130300 | + | 3045 | WP_001024766 | conjugal transfer protein | traU |
| HKK63_RS00925 | 130300..130596 | + | 297 | WP_012680982 | hypothetical protein | traV |
| HKK63_RS00930 | 130696..130920 | + | 225 | WP_284488195 | hypothetical protein | traV |
| HKK63_RS00675 (p026VIR_p145) | 130878..132083 | + | 1206 | WP_168989887 | conjugal transfer protein TraW | traW |
| HKK63_RS00680 (p026VIR_p146) | 132080..132649 | + | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| HKK63_RS00685 (p026VIR_p147) | 132724..134889 | + | 2166 | WP_168989888 | DotA/TraY family protein | traY |
| HKK63_RS00690 (p026VIR_p148) | 134977..135630 | + | 654 | WP_001402874 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HKK63_RS00695 (p026VIR_p149) | 135904..137490 | + | 1587 | WP_012680985 | TnsA endonuclease N-terminal domain-containing protein | - |
| HKK63_RS00700 (p026VIR_p150) | 137628..137780 | - | 153 | WP_001302699 | Hok/Gef family protein | - |
| HKK63_RS00705 (p026VIR_p151) | 138110..138352 | + | 243 | WP_001363144 | hypothetical protein | - |
| HKK63_RS00710 (p026VIR_p153) | 138652..139020 | + | 369 | WP_001363143 | hypothetical protein | - |
| HKK63_RS00715 (p026VIR_p154) | 139024..139239 | + | 216 | WP_000266543 | hypothetical protein | - |
| HKK63_RS00720 (p026VIR_p155) | 139437..140060 | - | 624 | WP_001155250 | helix-turn-helix domain-containing protein | - |
| HKK63_RS00725 (p026VIR_p156) | 140220..140687 | + | 468 | WP_000598962 | thermonuclease family protein | - |
| HKK63_RS00730 (p026VIR_p157) | 140837..141013 | + | 177 | WP_001054907 | hypothetical protein | - |
| HKK63_RS00935 | 141076..141171 | - | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HKK63_RS00940 | 141760..141855 | - | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HKK63_RS00740 (p026VIR_p162) | 142392..143708 | + | 1317 | WP_044192348 | IncI1-type conjugal transfer protein TrbA | trbA |
| HKK63_RS00745 (p026VIR_p163) | 143705..144829 | + | 1125 | WP_000152384 | DsbC family protein | trbB |
| HKK63_RS00750 (p026VIR_p164) | 144810..147110 | + | 2301 | WP_000077653 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 783 | GenBank | NC_012487 |
| Plasmid name | pO26-Vir | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 168100 bp | Coordinate of oriT [Strand] | 68493..68584 [+]; 150410..150495 [+] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | virulence region: toxB gene, the serine protease gene (espP), the catalase-peroxidase gene (katP), and the hemolysin (hly) gene cluster |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |