Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100290 |
| Name | oriT_pXFAS01 |
| Organism | Xylella fastidiosa M23 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_010579 (26824..26926 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 53..70, 75..92 (GTGAAGAAGGAACACCGG..CCGGTGGGCCTACTTCAC) |
| Location of nic site | 100..101 |
| Conserved sequence flanking the nic site |
TATCCTG|C |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 103 nt
>oriT_pXFAS01
AGCCTCGCAGAGCAGGATTCCCGTTGAGTACCGAAGGTGCGAATAAGGGAAAGTGAAGAAGGAACACCGGATTTCCGGTGGGCCTACTTCACATATCCTGCCC
AGCCTCGCAGAGCAGGATTCCCGTTGAGTACCGAAGGTGCGAATAAGGGAAAGTGAAGAAGGAACACCGGATTTCCGGTGGGCCTACTTCACATATCCTGCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file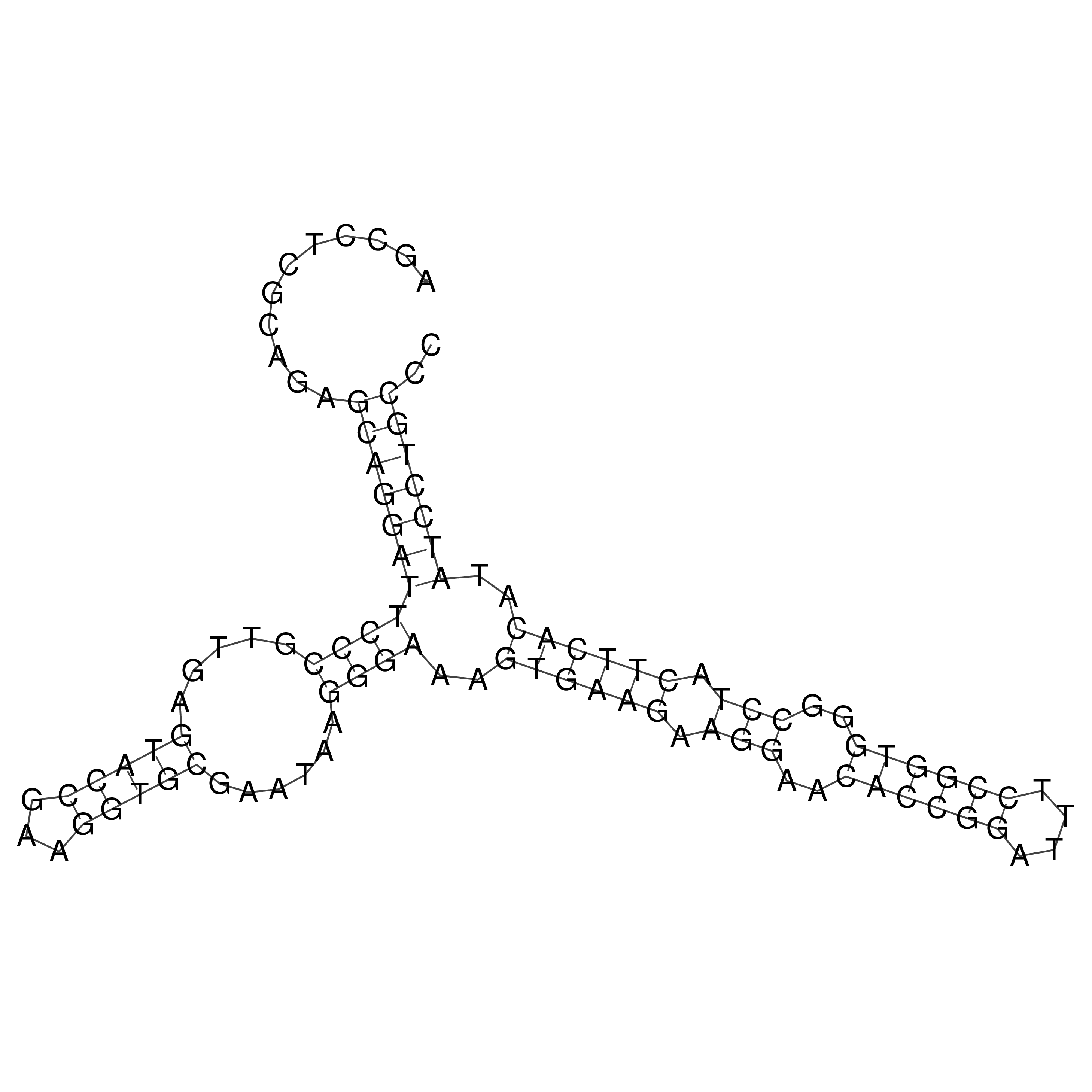
Relaxase
| ID | 361 | GenBank | WP_004086668 |
| Name | TrbI_pXFAS01 |
UniProt ID | B2IAU3 |
| Length | 830 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 830 a.a. Molecular weight: 92664.59 Da Isoelectric Point: 10.5711
>WP_004086668.1 TraI/MobA(P) family conjugative relaxase [Xylella fastidiosa]
MIAKHVPMRVVKKSDFRELVKYLSNQQGKQERVGCVTVTNCFQNNVLDAALEVQATQALNTRSEADKTYH
LLISFREGENPAPEILEVIESRVCAALGYADYQRVSVVHHDTDNLHIHVAINKIHPKSYTIHTPYNDYKT
LGEICKKLEREYGLEADNHTVRKTVGENRADDMERNAGVESLLGWIKRECTDQIKQVQSWSELHAVMQRN
GLEIRERGNGLVITNGVGRSVKASSVARDFSKAKLEARFGAFESFVKQAPSVSSVHARRIQAPLVEKVGR
RPPPRSKGRTPSLSSVGVLHVNSGIRYEQRPIYVKQASTAELYAMYKLEQQDLGAVRNVAIARARTKRDR
KIEATKRLGRIKRAAIKLMRGPGVNKKLLYALARKSLKEGVEKANTDYLKARDAAYATYHRRVWADWLQV
QATQGNAEALAALRAREMRQWWSCNIFGGLQARRTGPVPGLKPDNITKAGTIIYRVGSTAIRDDGDLLNV
SHGAGDYGVEAALRMAMYRYGERITVKGSDEFKKRVVQIAAAARLNISFDDEVLDKRRKQLVSDAAVIVK
LTDAMLSDKTAQFTQAQASRVGAFQEDALSEYDAAISRSINLANQEQINEQQRTRAKHYFVQRWSDRGGP
NSRRDGHATRIRSGGGGSGKHGASGATVISSAGGFNKPNVGGIGAKPPPASTHRLRNLHQLGVVQFSRGS
EVLLPRHVSDCVGEQQTERNHRVRRDVHTAGITSSELMAVDKYIAERELKRVREFDIMKNRRYNEDDAGM
VKFAGLRQVDGKSLVLLKRNDEVIVLAIDVATARRMKRFSLGDEVLLTKKGTLKTKGRNR
MIAKHVPMRVVKKSDFRELVKYLSNQQGKQERVGCVTVTNCFQNNVLDAALEVQATQALNTRSEADKTYH
LLISFREGENPAPEILEVIESRVCAALGYADYQRVSVVHHDTDNLHIHVAINKIHPKSYTIHTPYNDYKT
LGEICKKLEREYGLEADNHTVRKTVGENRADDMERNAGVESLLGWIKRECTDQIKQVQSWSELHAVMQRN
GLEIRERGNGLVITNGVGRSVKASSVARDFSKAKLEARFGAFESFVKQAPSVSSVHARRIQAPLVEKVGR
RPPPRSKGRTPSLSSVGVLHVNSGIRYEQRPIYVKQASTAELYAMYKLEQQDLGAVRNVAIARARTKRDR
KIEATKRLGRIKRAAIKLMRGPGVNKKLLYALARKSLKEGVEKANTDYLKARDAAYATYHRRVWADWLQV
QATQGNAEALAALRAREMRQWWSCNIFGGLQARRTGPVPGLKPDNITKAGTIIYRVGSTAIRDDGDLLNV
SHGAGDYGVEAALRMAMYRYGERITVKGSDEFKKRVVQIAAAARLNISFDDEVLDKRRKQLVSDAAVIVK
LTDAMLSDKTAQFTQAQASRVGAFQEDALSEYDAAISRSINLANQEQINEQQRTRAKHYFVQRWSDRGGP
NSRRDGHATRIRSGGGGSGKHGASGATVISSAGGFNKPNVGGIGAKPPPASTHRLRNLHQLGVVQFSRGS
EVLLPRHVSDCVGEQQTERNHRVRRDVHTAGITSSELMAVDKYIAERELKRVREFDIMKNRRYNEDDAGM
VKFAGLRQVDGKSLVLLKRNDEVIVLAIDVATARRMKRFSLGDEVLLTKKGTLKTKGRNR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B2IAU3 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3..8817
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| XFASM23_RS11300 (XfasM23_2227) | 3..323 | + | 321 | WP_004086725 | conjugal transfer protein TrbD | virB3 |
| XFASM23_RS11305 (XfasM23_2228) | 311..2875 | + | 2565 | WP_004086690 | conjugal transfer protein TrbE | virb4 |
| XFASM23_RS11310 (XfasM23_2229) | 2872..3588 | + | 717 | WP_004086692 | conjugal transfer protein TrbF | virB8 |
| XFASM23_RS11315 (XfasM23_2230) | 3605..4495 | + | 891 | WP_004086695 | P-type conjugative transfer protein TrbG | virB9 |
| XFASM23_RS11320 (XfasM23_2231) | 4498..4974 | + | 477 | WP_004086697 | conjugal transfer protein TrbH | - |
| XFASM23_RS11325 (XfasM23_2232) | 4980..6380 | + | 1401 | WP_012382833 | TrbI/VirB10 family protein | virB10 |
| XFASM23_RS11330 (XfasM23_2233) | 6399..7172 | + | 774 | WP_004086702 | P-type conjugative transfer protein TrbJ | virB5 |
| XFASM23_RS11335 (XfasM23_2234) | 7189..7425 | + | 237 | WP_012382834 | EexN family lipoprotein | - |
| XFASM23_RS11340 (XfasM23_2235) | 7447..8817 | + | 1371 | WP_004086703 | P-type conjugative transfer protein TrbL | virB6 |
| XFASM23_RS11345 (XfasM23_2236) | 8823..9422 | + | 600 | WP_004086719 | conjugal transfer protein TrbN | - |
| XFASM23_RS11355 (XfasM23_2237) | 10035..10262 | - | 228 | WP_004086706 | hypothetical protein | - |
| XFASM23_RS11360 (XfasM23_2238) | 10259..10627 | - | 369 | WP_004086708 | hypothetical protein | - |
| XFASM23_RS11365 (XfasM23_2239) | 11170..11733 | + | 564 | WP_004086710 | recombinase family protein | - |
| XFASM23_RS11370 (XfasM23_2240) | 12064..12657 | + | 594 | WP_004086726 | LPD29 domain-containing protein | - |
| XFASM23_RS11375 (XfasM23_2241) | 12725..13039 | + | 315 | WP_012382835 | TrfB-related DNA-binding protein | - |
| XFASM23_RS11380 (XfasM23_2242) | 13216..13809 | + | 594 | WP_004086728 | LPD29 domain-containing protein | - |
Host bacterium
| ID | 752 | GenBank | NC_010579 |
| Plasmid name | pXFAS01 | Incompatibility group | _ |
| Plasmid size | 38297 bp | Coordinate of oriT [Strand] | 26824..26926 [+] |
| Host baterium | Xylella fastidiosa M23 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |