Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100260 |
| Name | oriT_pJMP134-1 |
| Organism | Cupriavidus pinatubonensis JMP134 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_007337 (85714..86086 [+], 373 nt) |
| oriT length | 373 nt |
| IRs (inverted repeats) | IR1: 64..68, 72..76 (GCGCC..GGCGC) IR2: 89..107, 110..128 (GTGAAGATAGATAACCGGC..GCCGGTTAGCTAACTTCAC) |
| Location of nic site | 136..137 |
| Conserved sequence flanking the nic site |
CATCCTG|C |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 373 nt
>oriT_pJMP134-1
GCCTCGCCTCGTCTCTCATGCCGGCGGTAGCCGGCTGCCTCGCAGAGCAGGATGACCCGTTGAGCGCCCCCGGCGCGAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACACCAGCCATTACGATTTATCCGCAACTATCGCGCTATCAGGCCGCAAAAGCAGCAACGGATATAGCGAAAACCGCCACAATGGCCCATAATGCCGCTATCGAAGCGTGCCAATGCACGCCGATAGCGGACTTTTTGCGTTTCCGTAGCGCCGCTTAGTAGCGTTACATTTGCGATGAGAGGATTAGATGGACGAACACG
GCCTCGCCTCGTCTCTCATGCCGGCGGTAGCCGGCTGCCTCGCAGAGCAGGATGACCCGTTGAGCGCCCCCGGCGCGAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACACCAGCCATTACGATTTATCCGCAACTATCGCGCTATCAGGCCGCAAAAGCAGCAACGGATATAGCGAAAACCGCCACAATGGCCCATAATGCCGCTATCGAAGCGTGCCAATGCACGCCGATAGCGGACTTTTTGCGTTTCCGTAGCGCCGCTTAGTAGCGTTACATTTGCGATGAGAGGATTAGATGGACGAACACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file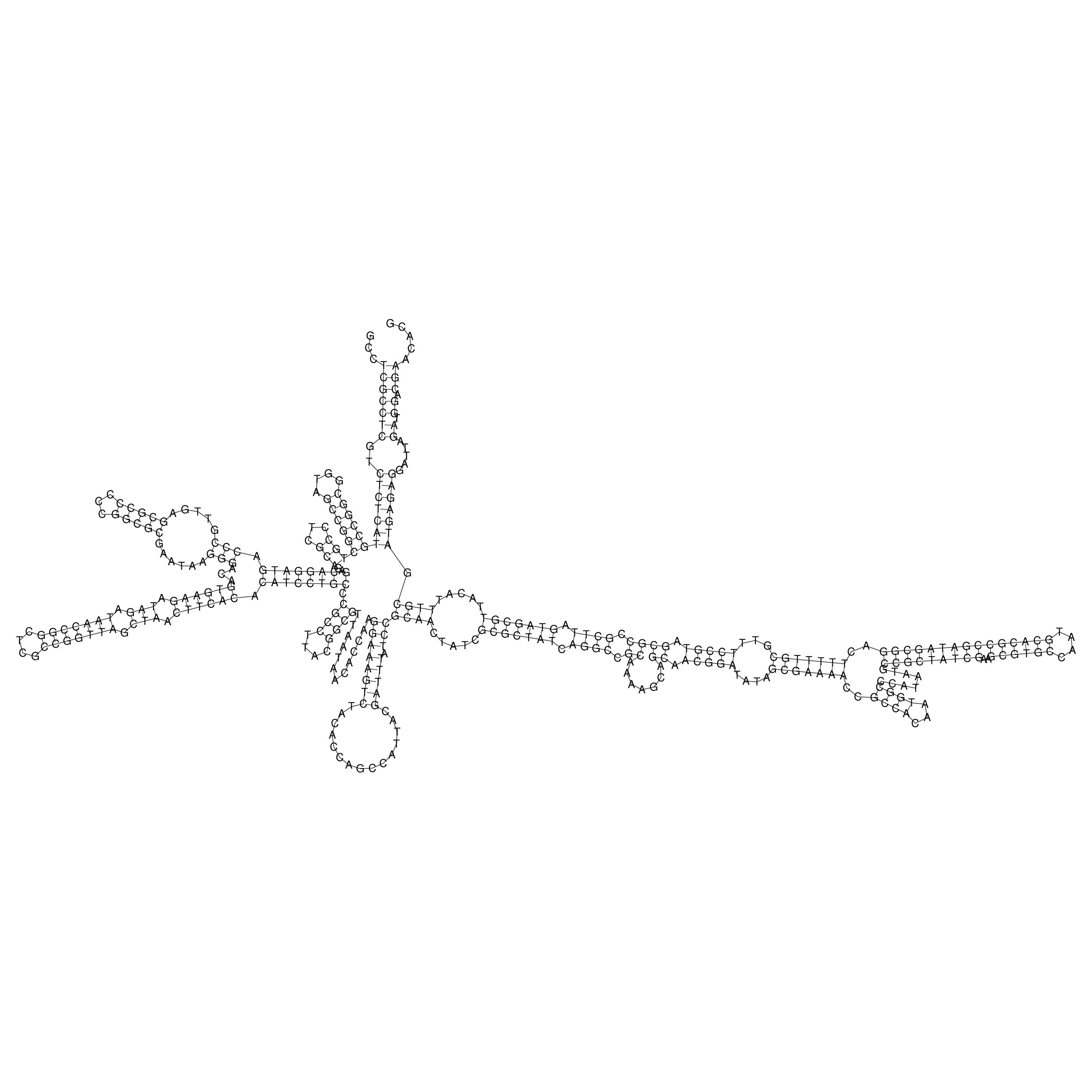
Relaxase
| ID | 331 | GenBank | WP_011114057 |
| Name | TraI_pJMP134-1 |
UniProt ID | Q6UP38 |
| Length | 746 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82038.71 Da Isoelectric Point: 10.7900
>WP_011114057.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Bacteria]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q6UP38 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 58243..68920
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| REUT_RS32975 (Reut_D6496) | 54470..54745 | - | 276 | WP_003131987 | mercury resistance system periplasmic binding protein MerP | - |
| REUT_RS32980 (Reut_D6497) | 54758..55108 | - | 351 | WP_003131974 | mercuric ion transporter MerT | - |
| REUT_RS32985 (Reut_D6498) | 55180..55614 | + | 435 | WP_003131969 | mercury resistance transcriptional regulator MerR | - |
| REUT_RS32990 (Reut_D6499) | 55727..56947 | - | 1221 | WP_011114072 | plasmid replication initiator TrfA | - |
| REUT_RS32995 (Reut_D6500) | 56994..57335 | - | 342 | WP_000019167 | single-stranded DNA-binding protein | - |
| REUT_RS33000 (Reut_D6501) | 57571..57933 | + | 363 | WP_011114045 | transcriptional regulator | - |
| REUT_RS33005 (Reut_D6502) | 58243..59205 | + | 963 | WP_000132020 | P-type conjugative transfer ATPase TrbB | virB11 |
| REUT_RS33010 (Reut_D6503) | 59222..59686 | + | 465 | WP_011114046 | conjugal transfer system pilin TrbC | virB2 |
| REUT_RS33015 (Reut_D6504) | 59690..60001 | + | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| REUT_RS33020 (Reut_D6505) | 59998..62556 | + | 2559 | WP_011114047 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| REUT_RS33025 (Reut_D6506) | 62553..63335 | + | 783 | WP_011114048 | conjugal transfer protein TrbF | virB8 |
| REUT_RS33030 (Reut_D6507) | 63353..64252 | + | 900 | WP_006122480 | P-type conjugative transfer protein TrbG | virB9 |
| REUT_RS33035 (Reut_D6508) | 64255..64743 | + | 489 | WP_006122479 | conjugal transfer protein TrbH | - |
| REUT_RS33040 (Reut_D6509) | 64748..66169 | + | 1422 | WP_011114050 | TrbI/VirB10 family protein | virB10 |
| REUT_RS33045 (Reut_D6510) | 66190..66954 | + | 765 | WP_010890135 | P-type conjugative transfer protein TrbJ | virB5 |
| REUT_RS33050 (Reut_D6511) | 66964..67191 | + | 228 | WP_000644148 | entry exclusion lipoprotein TrbK | - |
| REUT_RS33055 (Reut_D6512) | 67202..68920 | + | 1719 | WP_011114051 | P-type conjugative transfer protein TrbL | virB6 |
| REUT_RS33060 (Reut_D6513) | 68938..69525 | + | 588 | WP_010890138 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| REUT_RS33065 (Reut_D6514) | 69539..70174 | + | 636 | WP_011114052 | transglycosylase SLT domain-containing protein | - |
| REUT_RS33070 (Reut_D6515) | 70203..70469 | + | 267 | WP_010890140 | hypothetical protein | - |
| REUT_RS33075 (Reut_D6516) | 70469..71167 | + | 699 | WP_011114053 | TraX family protein | - |
| REUT_RS33080 (Reut_D6517) | 71183..71614 | + | 432 | WP_001665065 | OmpA family protein | - |
| REUT_RS33085 (Reut_D6518) | 71769..72443 | + | 675 | WP_011178411 | DNA methylase | - |
| REUT_RS33090 (Reut_D6519) | 72445..73104 | - | 660 | WP_011178412 | recombinase family protein | - |
Host bacterium
| ID | 722 | GenBank | NC_007337 |
| Plasmid name | pJMP134-1 | Incompatibility group | IncP1 |
| Plasmid size | 87688 bp | Coordinate of oriT [Strand] | 85714..86086 [+] |
| Host baterium | Cupriavidus pinatubonensis JMP134 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | mercury resistance |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |