Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100245 |
| Name | oriT_pPSR1 |
| Organism | Pseudomonas syringae pv. syringae |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_005205 (66771..66916 [+], 146 nt) |
| oriT length | 146 nt |
| IRs (inverted repeats) | IR1: 38..43, 52..57 (AAAAAG..CTTTTT) IR2: 72..83, 87..98 (GTGCCAAAGGGG..CCCCTTTGAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 146 nt
>oriT_pPSR1
CGAATTCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTATCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCGGAGCGGGAAAGCGTTGTCGTTGTCGTTGTTGGTTGTTGTCAT
CGAATTCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTATCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCGGAGCGGGAAAGCGTTGTCGTTGTCGTTGTTGGTTGTTGTCAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file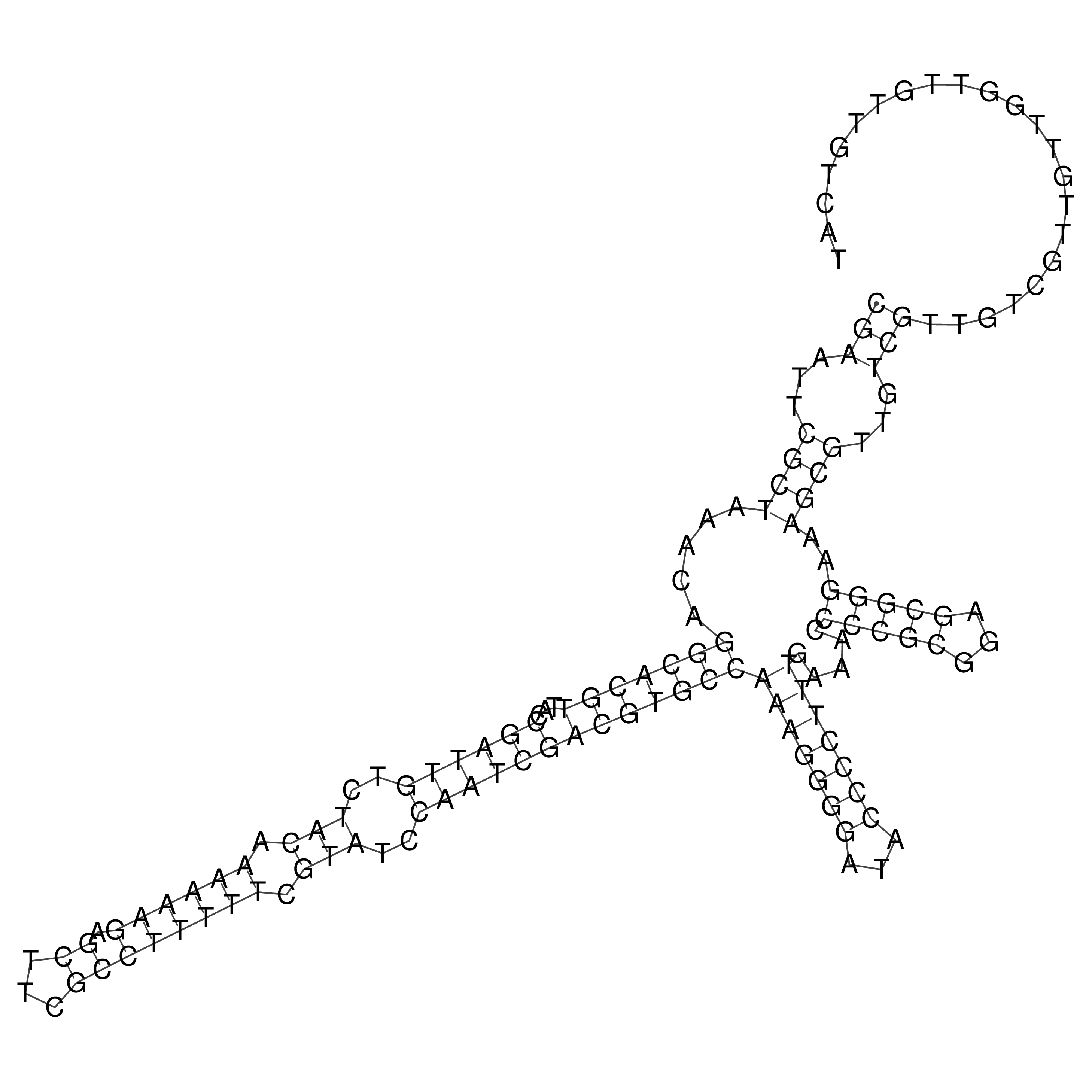
Relaxase
| ID | 316 | GenBank | NP_940739 |
| Name | Orf54_pPSR1 |
UniProt ID | Q6VE73 |
| Length | 1249 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1249 a.a. Molecular weight: 138583.89 Da Isoelectric Point: 8.9877
>NP_940739.1 Orf54 (plasmid) [Pseudomonas syringae pv. syringae]
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDDFIVRRELRKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERETNFYRAHGEDYETVHTGQRQRDHQRSPA
ETTGRRTAAPADGVQDLSVSDVADHRQAGPTRSRDGALLLPSDAHVHVGQSQPGGDSGLRSSVPGGGRGR
RAGSTAERGRGGRQPAAVSQETKGTATPAGATGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSHPQVADQLPQARAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKVLVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPGPEGVRERVKRVMDRFAKQVDPVAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKSMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADRLGATVPIEALYGQGKTADQVSQALTAQL
DTVPESERIAFVETVAITLGIPERGQPKGDEAFAQWQAQRAQPAANSVASATSEAQANVPTPSDAVPEPV
NPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMGLRGEDHAIWIIATDDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPQSVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIDHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEEAMSDADLKQGDQVRLQDLGTQPVVV
QVLEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAAAAGTPELSSPDEEDGMNMD
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDDFIVRRELRKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERETNFYRAHGEDYETVHTGQRQRDHQRSPA
ETTGRRTAAPADGVQDLSVSDVADHRQAGPTRSRDGALLLPSDAHVHVGQSQPGGDSGLRSSVPGGGRGR
RAGSTAERGRGGRQPAAVSQETKGTATPAGATGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSHPQVADQLPQARAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKVLVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPGPEGVRERVKRVMDRFAKQVDPVAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKSMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADRLGATVPIEALYGQGKTADQVSQALTAQL
DTVPESERIAFVETVAITLGIPERGQPKGDEAFAQWQAQRAQPAANSVASATSEAQANVPTPSDAVPEPV
NPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEADVRASDTVMGLRGEDHAIWIIATDDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPQSVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIDHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEEAMSDADLKQGDQVRLQDLGTQPVVV
QVLEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAAAAGTPELSSPDEEDGMNMD
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q6VE73 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 47681..58028
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS292_RS00225 | 43033..43263 | - | 231 | Protein_48 | transposase domain-containing protein | - |
| HS292_RS00230 | 43490..43828 | + | 339 | Protein_49 | tyrosine-type recombinase/integrase | - |
| HS292_RS00235 | 43989..44471 | + | 483 | WP_054081655 | hypothetical protein | - |
| HS292_RS00240 | 45047..45433 | - | 387 | WP_228409379 | hypothetical protein | - |
| HS292_RS00245 (pPSR1_p35) | 45673..46143 | - | 471 | WP_011152902 | DUF6392 family protein | - |
| HS292_RS00250 (pPSR1_p36) | 46230..46502 | - | 273 | WP_230081992 | helix-turn-helix transcriptional regulator | - |
| HS292_RS00255 (pPSR1_p37) | 46844..47371 | + | 528 | WP_011152904 | transcription termination/antitermination NusG family protein | - |
| HS292_RS00260 | 47443..47652 | + | 210 | WP_223812427 | hypothetical protein | - |
| HS292_RS00265 (pPSR1_p38) | 47681..48388 | + | 708 | WP_011152905 | lytic transglycosylase domain-containing protein | virB1 |
| HS292_RS00270 (pPSR1_p39) | 48388..48723 | + | 336 | WP_173479888 | conjugal transfer protein | - |
| HS292_RS00275 (pPSR1_p40) | 48736..49224 | + | 489 | WP_011152907 | VirB3 family type IV secretion system protein | virB3 |
| HS292_RS00280 (pPSR1_p41) | 49127..51619 | + | 2493 | WP_228409383 | conjugal transfer protein | virb4 |
| HS292_RS00285 (pPSR1_p42) | 51622..52308 | + | 687 | WP_011152909 | P-type DNA transfer protein VirB5 | - |
| HS292_RS00290 | 52342..52695 | + | 354 | WP_054081656 | hypothetical protein | - |
| HS292_RS00295 (pPSR1_p43) | 52706..53638 | + | 933 | WP_011152910 | type IV secretion system protein | virB6 |
| HS292_RS00300 (pPSR1_p45) | 54046..54834 | + | 789 | WP_011152912 | type IV secretion system protein | virB8 |
| HS292_RS00305 (pPSR1_p46) | 54824..55633 | + | 810 | WP_011152913 | P-type conjugative transfer protein VirB9 | virB9 |
| HS292_RS00310 (pPSR1_p47) | 55620..56987 | + | 1368 | WP_011152914 | TrbI/VirB10 family protein | virB10 |
| HS292_RS00315 (pPSR1_p48) | 56997..58028 | + | 1032 | WP_011152915 | P-type DNA transfer ATPase VirB11 | virB11 |
| HS292_RS00320 | 58038..58394 | + | 357 | WP_010217013 | hypothetical protein | - |
| HS292_RS00325 | 58584..58814 | + | 231 | WP_054081657 | hypothetical protein | - |
| HS292_RS00330 (pPSR1_p49) | 58814..60472 | + | 1659 | WP_011152916 | type IV secretory system conjugative DNA transfer family protein | - |
| HS292_RS00335 | 60513..60875 | + | 363 | WP_054081658 | TrbM/KikA/MpfK family conjugal transfer protein | - |
Host bacterium
| ID | 707 | GenBank | NC_005205 |
| Plasmid name | pPSR1 | Incompatibility group | _ |
| Plasmid size | 72601 bp | Coordinate of oriT [Strand] | 66771..66916 [+] |
| Host baterium | Pseudomonas syringae pv. syringae |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |