Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100234 |
| Name | oriT_pCTX-M3 |
| Organism | Citrobacter freundii |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_004464 (31616..31721 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 18..35, 41..58 (GTTTTTGGAGTACCGCCG..CGGCGGCCGTCCAAAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
ACATCTTGT |
| Note |
oriT sequence
Download Length: 106 nt
>oriT_pCTX-M3
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file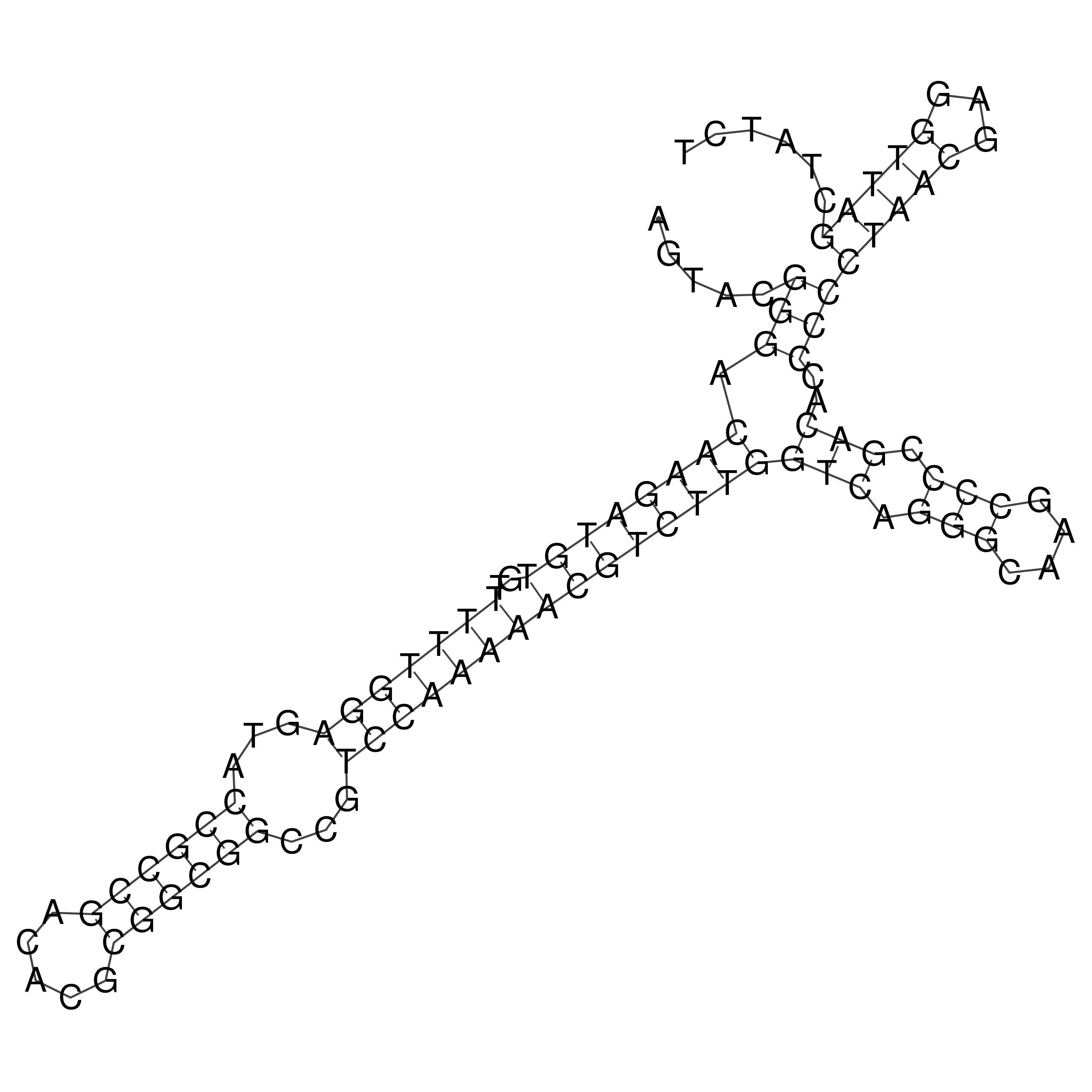
Reference
[1] Michał Dmowski et al. (2018) Characteristics of the Conjugative Transfer System of the IncM Plasmid pCTX-M3 and Identification of Its Putative Regulators. Journal of bacteriology. 200(18):e00234-18. [PMID:29986941]
Relaxase
| ID | 305 | GenBank | NP_775011 |
| Name | NikB_pCTX-M3 |
UniProt ID | Q8GFT3 |
| Length | 658 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 658 a.a. Molecular weight: 75135.84 Da Isoelectric Point: 10.0285
>NP_775011.1 putative nickase (plasmid) [Citrobacter freundii]
MIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVDV
EPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLAS
MGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVNDR
QQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHKT
FADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTAY
TPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPVYSPDKDRIADEYRKIAQHTRLVKNNVRHSVS
DPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSSR
KKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLTE
SNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQNE
KHRLIREQNNSKNEMQFRSERDDKFNPK
MIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVDV
EPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLAS
MGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVNDR
QQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHKT
FADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTAY
TPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPVYSPDKDRIADEYRKIAQHTRLVKNNVRHSVS
DPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSSR
KKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLTE
SNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQNE
KHRLIREQNNSKNEMQFRSERDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q8GFT3 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 34517..53620
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKK50_RS00250 (pCTX-M3_048) | 29601..29912 | + | 312 | WP_011091065 | hypothetical protein | - |
| HKK50_RS00255 (pCTX-M3_049) | 30045..30581 | + | 537 | WP_011091066 | hypothetical protein | - |
| HKK50_RS00260 (pCTX-M3_050) | 31082..31447 | - | 366 | WP_011091067 | hypothetical protein | - |
| HKK50_RS00550 | 31462..32040 | + | 579 | WP_223811519 | plasmid mobilization protein MobA | - |
| HKK50_RS00270 (pCTX-M3_052) | 32027..34006 | + | 1980 | WP_050868819 | TraI/MobA(P) family conjugative relaxase | - |
| HKK50_RS00275 (pCTX-M3_053) | 34020..34520 | + | 501 | WP_004187320 | DotD/TraH family lipoprotein | - |
| HKK50_RS00280 (pCTX-M3_054) | 34517..35296 | + | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| HKK50_RS00285 (pCTX-M3_055) | 35307..36470 | + | 1164 | WP_004187313 | plasmid transfer ATPase TraJ | virB11 |
| HKK50_RS00290 (pCTX-M3_056) | 36460..36720 | + | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| HKK50_RS00555 | 36745..39957 | + | 3213 | WP_011091070 | LPD7 domain-containing protein | - |
| HKK50_RS00300 (pCTX-M3_058) | 39881..40435 | + | 555 | WP_011154467 | hypothetical protein | traL |
| HKK50_RS00305 | 40436..40633 | - | 198 | WP_004187464 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| HKK50_RS00310 | 40620..41030 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| HKK50_RS00315 (pCTX-M3_060) | 41029..41811 | + | 783 | WP_011091073 | DotI/IcmL family type IV secretion protein | traM |
| HKK50_RS00320 (pCTX-M3_061) | 41820..42971 | + | 1152 | WP_011091074 | DotH/IcmK family type IV secretion protein | traN |
| HKK50_RS00325 (pCTX-M3_062) | 42983..44332 | + | 1350 | WP_011091075 | conjugal transfer protein TraO | traO |
| HKK50_RS00330 (pCTX-M3_063) | 44344..45048 | + | 705 | WP_011091076 | conjugal transfer protein TraP | traP |
| HKK50_RS00335 (pCTX-M3_064) | 45072..45602 | + | 531 | WP_011091077 | conjugal transfer protein TraQ | traQ |
| HKK50_RS00340 (pCTX-M3_065) | 45619..46008 | + | 390 | WP_011091078 | DUF6750 family protein | traR |
| HKK50_RS00345 (pCTX-M3_067) | 46054..46548 | + | 495 | WP_011091080 | hypothetical protein | - |
| HKK50_RS00350 (pCTX-M3_068) | 46545..49595 | + | 3051 | WP_011091081 | hypothetical protein | traU |
| HKK50_RS00355 (pCTX-M3_069) | 49592..50800 | + | 1209 | WP_011091082 | conjugal transfer protein TraW | traW |
| HKK50_RS00360 (pCTX-M3_070) | 50797..51447 | + | 651 | WP_011091083 | hypothetical protein | - |
| HKK50_RS00365 (pCTX-M3_071) | 51440..53620 | + | 2181 | WP_004187492 | DotA/TraY family protein | traY |
| HKK50_RS00370 (pCTX-M3_072) | 53623..54276 | + | 654 | WP_011091084 | hypothetical protein | - |
| HKK50_RS00375 (pCTX-M3_073) | 54333..54581 | + | 249 | WP_015062836 | IncL/M type plasmid replication protein RepC | - |
| HKK50_RS00380 (pCTX-M3_075) | 54867..55934 | + | 1068 | WP_050868820 | plasmid replication initiator RepA | - |
| HKK50_RS00385 (pCTX-M3_076) | 56938..57798 | - | 861 | WP_000027057 | broad-spectrum class A beta-lactamase TEM-1 | - |
Host bacterium
| ID | 696 | GenBank | NC_004464 |
| Plasmid name | pCTX-M3 | Incompatibility group | IncL/M |
| Plasmid size | 89468 bp | Coordinate of oriT [Strand] | 31616..31721 [+] |
| Host baterium | Citrobacter freundii |
Cargo genes
| Drug resistance gene | blaCTX-M-3, blaTEM-1B, aac(3)-IId, dfrA12, aadA2, qacE, sul1, armA, msr(E), mph(E) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |