Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100224 |
| Name | oriT_Ti |
| Organism | Agrobacterium tumefaciens |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_002377 (30720..30756 [+], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 18..23, 27..32 (CGTCGT..ACGACG) |
| Location of nic site | 7..8 |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 37 nt
>oriT_Ti
CCAAGGGCGCAATTATACGTCGTGATACGACGCGTTG
CCAAGGGCGCAATTATACGTCGTGATACGACGCGTTG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file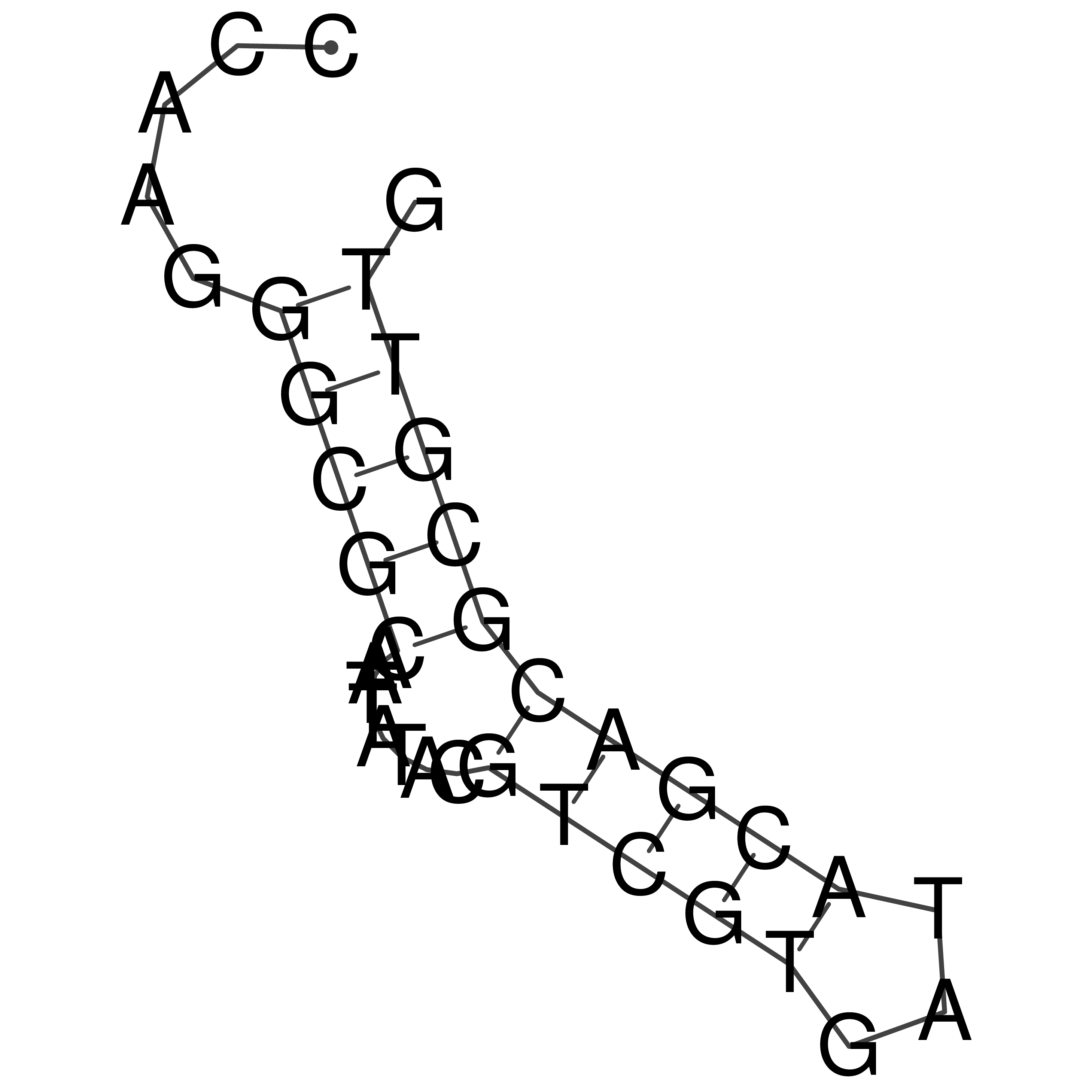
Relaxase
| ID | 295 | GenBank | NP_059695 |
| Name | TraA_Ti |
UniProt ID | Q44363 |
| Length | 1100 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123724.31 Da Isoelectric Point: 10.0043
>NP_059695.1 hypothetical protein pTi_023 (plasmid) [Agrobacterium tumefaciens]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRSLIADR
SVSGASEAFWNKVEAFEKRADAQLARDLTIALPLELSAEQNIALVRDFVENHILAKGMVADWVYHENPGN
PHIHLMTTLRPLSDESFGSKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGINLEPTIHLGVGAKAIERKAEQRGVRPELERVELNEQRRSENTRRILNNPAIVLDL
ITREKSVFDERDVAKVLHRYIDDPALFQQLMIKIILNRQVLRLQRETIDFSTGEKLPARYSTRAMIRLEA
TMARQATWLSNREGRGVSPTALDATFRRHERLSDEQKAAIEHVAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYHVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKRGRDLLDDKTIFIMDEAGMVASKQMAGFVD
TAVRAGAKIVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREEWMRKASLDLARGNVENALSAYR
ANVRITGERLKAEAVERLIADWNHDYDQTKTILILAHLRRDVRMLNVMAREKLVERGMVGEGHLFRTADG
ERRFDAGDQIVFLKNEGSLGLKNGMIGHVVVAAANRIVATVGEGDQRRQVIVEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRRQAKETTLDYEHGQFY
REALRFAETRGLHVVQVARTMVRDRLDWTLRQKAKLVDLSRRLAAFAAHLGITQSPKTQTMKETAPMVAG
IKTFSGSVSDIVGDRLGTDPSLKRQWEEVSARFAYVFADPETAFRAMNFAALLADKEVAKQILQKLEAEP
GSIGPLKGKTGILASKPEREARRVAEVNVPALKRDLEQYLRMRETVTQRLQTDEQSLRQRVSIDIPALSP
AARVVRERVRDAIDRNDLPAAIAYALSNRETKLEIDGFNQAVTERFGERTLLSNAAREPSGKLYEKLSEG
MKPEQKEELKQAWPVMRTAQQLVAHERTVHSLKLAEEHRLTQRQTPVLKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRSLIADR
SVSGASEAFWNKVEAFEKRADAQLARDLTIALPLELSAEQNIALVRDFVENHILAKGMVADWVYHENPGN
PHIHLMTTLRPLSDESFGSKKVAVIGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGINLEPTIHLGVGAKAIERKAEQRGVRPELERVELNEQRRSENTRRILNNPAIVLDL
ITREKSVFDERDVAKVLHRYIDDPALFQQLMIKIILNRQVLRLQRETIDFSTGEKLPARYSTRAMIRLEA
TMARQATWLSNREGRGVSPTALDATFRRHERLSDEQKAAIEHVAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYHVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKRGRDLLDDKTIFIMDEAGMVASKQMAGFVD
TAVRAGAKIVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREEWMRKASLDLARGNVENALSAYR
ANVRITGERLKAEAVERLIADWNHDYDQTKTILILAHLRRDVRMLNVMAREKLVERGMVGEGHLFRTADG
ERRFDAGDQIVFLKNEGSLGLKNGMIGHVVVAAANRIVATVGEGDQRRQVIVEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRRQAKETTLDYEHGQFY
REALRFAETRGLHVVQVARTMVRDRLDWTLRQKAKLVDLSRRLAAFAAHLGITQSPKTQTMKETAPMVAG
IKTFSGSVSDIVGDRLGTDPSLKRQWEEVSARFAYVFADPETAFRAMNFAALLADKEVAKQILQKLEAEP
GSIGPLKGKTGILASKPEREARRVAEVNVPALKRDLEQYLRMRETVTQRLQTDEQSLRQRVSIDIPALSP
AARVVRERVRDAIDRNDLPAAIAYALSNRETKLEIDGFNQAVTERFGERTLLSNAAREPSGKLYEKLSEG
MKPEQKEELKQAWPVMRTAQQLVAHERTVHSLKLAEEHRLTQRQTPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q44363 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 96666..106317
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKJ77_RS00430 (pTi_075) | 92102..92983 | + | 882 | WP_010892435 | ABC transporter permease | - |
| HKJ77_RS00435 (pTi_076) | 92983..94749 | + | 1767 | WP_010892436 | ABC transporter ATP-binding protein | - |
| HKJ77_RS00440 (pTi_077) | 94901..96433 | + | 1533 | WP_323130362 | ABC transporter substrate-binding protein | - |
| HKJ77_RS00445 (pTi_078) | 96666..97967 | - | 1302 | WP_010892438 | IncP-type conjugal transfer protein TrbI | virB10 |
| HKJ77_RS00450 (pTi_079) | 97977..98453 | - | 477 | WP_046205713 | conjugal transfer protein TrbH | - |
| HKJ77_RS00455 (pTi_080) | 98453..99307 | - | 855 | WP_010892440 | P-type conjugative transfer protein TrbG | virB9 |
| HKJ77_RS00460 (pTi_081) | 99324..99986 | - | 663 | WP_010892441 | conjugal transfer protein TrbF | virB8 |
| HKJ77_RS00465 (pTi_082) | 100001..101188 | - | 1188 | WP_010892442 | P-type conjugative transfer protein TrbL | virB6 |
| HKJ77_RS00470 (pTi_083) | 101182..101409 | - | 228 | WP_010892443 | entry exclusion protein TrbK | - |
| HKJ77_RS00475 (pTi_084) | 101406..102215 | - | 810 | WP_010892444 | P-type conjugative transfer protein TrbJ | virB5 |
| HKJ77_RS00480 (pTi_085) | 102187..104649 | - | 2463 | WP_010892445 | conjugal transfer protein TrbE | virb4 |
| HKJ77_RS00485 (pTi_086) | 104660..104959 | - | 300 | WP_010892446 | conjugal transfer protein TrbD | virB3 |
| HKJ77_RS00490 (pTi_087) | 104952..105356 | - | 405 | WP_010892447 | conjugal transfer pilin TrbC | virB2 |
| HKJ77_RS00495 (pTi_088) | 105346..106317 | - | 972 | WP_010892448 | P-type conjugative transfer ATPase TrbB | virB11 |
| HKJ77_RS00500 (pTi_089) | 106314..106952 | - | 639 | WP_010892449 | acyl-homoserine-lactone synthase | - |
| HKJ77_RS00505 (pTi_090) | 107313..108530 | + | 1218 | WP_010892450 | plasmid partitioning protein RepA | - |
| HKJ77_RS00510 (pTi_091) | 108760..109770 | + | 1011 | WP_040132353 | plasmid partitioning protein RepB | - |
| HKJ77_RS00515 (pTi_092) | 109976..111295 | + | 1320 | WP_010892452 | plasmid replication protein RepC | - |
Region 2: 151232..160667
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HKJ77_RS00695 (pTi_126) | 148762..149529 | + | 768 | WP_010892486 | AcvB/VirJ family lysyl-phosphatidylglycerol hydrolase | - |
| HKJ77_RS00700 | 150091..150855 | + | 765 | Protein_143 | IS5 family transposase | - |
| HKJ77_RS00705 (pTi_127) | 151232..151951 | + | 720 | WP_010892487 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| HKJ77_RS00710 (pTi_128) | 152023..152331 | + | 309 | WP_233278929 | pilin major subunit VirB2 | virB2 |
| HKJ77_RS00715 (pTi_129) | 152331..152657 | + | 327 | WP_010892489 | type IV secretion system protein VirB3 | virB3 |
| HKJ77_RS00720 (pTi_130) | 152657..155026 | + | 2370 | WP_010892490 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HKJ77_RS00725 (pTi_131) | 155041..155703 | + | 663 | WP_010892491 | pilin minor subunit VirB5 | - |
| HKJ77_RS00730 (pTi_132) | 155800..156687 | + | 888 | WP_010892492 | type IV secretion system protein | virB6 |
| HKJ77_RS00735 (pTi_133) | 156721..156888 | + | 168 | WP_010892493 | type IV secretion system lipoprotein VirB7 | - |
| HKJ77_RS00740 (pTi_134) | 156875..157588 | + | 714 | WP_032489857 | type IV secretion system protein VirB8 | virB8 |
| HKJ77_RS00745 (pTi_135) | 157585..158466 | + | 882 | WP_010892495 | P-type conjugative transfer protein VirB9 | virB9 |
| HKJ77_RS00750 (pTi_136) | 158463..159596 | + | 1134 | WP_010892496 | type IV secretion system protein VirB10 | virB10 |
| HKJ77_RS00755 (pTi_137) | 159636..160667 | + | 1032 | WP_010892497 | P-type DNA transfer ATPase VirB11 | virB11 |
| HKJ77_RS00760 (pTi_138) | 160799..161602 | + | 804 | WP_032488325 | response regulator | - |
| HKJ77_RS00765 | 161661..162044 | + | 384 | WP_153463909 | hypothetical protein | - |
| HKJ77_RS00770 | 162100..162399 | - | 300 | Protein_157 | transposase domain-containing protein | - |
| HKJ77_RS00775 | 162427..164082 | - | 1656 | WP_046205714 | IS66-like element IS66 family transposase | - |
| HKJ77_RS00780 | 164128..164475 | - | 348 | WP_040132297 | IS66 family insertion sequence element accessory protein TnpB | - |
| HKJ77_RS00785 | 164472..164879 | - | 408 | WP_040132299 | transposase | - |
Host bacterium
| ID | 686 | GenBank | NC_002377 |
| Plasmid name | Ti | Incompatibility group | _ |
| Plasmid size | 194140 bp | Coordinate of oriT [Strand] | 30720..30756 [+] |
| Host baterium | Agrobacterium tumefaciens |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |