Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100218 |
| Name | oriT_R7K |
| Organism | Providencia rettgeri |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_010643 (9136..9174 [-], 39 nt) |
| oriT length | 39 nt |
| IRs (inverted repeats) | 9..14, 19..24 (CGCACC..GGTGCG) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTCT|ATAGC |
| Note | _ |
oriT sequence
Download Length: 39 nt
GACTTACGCGCACCGAAAGGTGCGTATTGTCTATAGCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file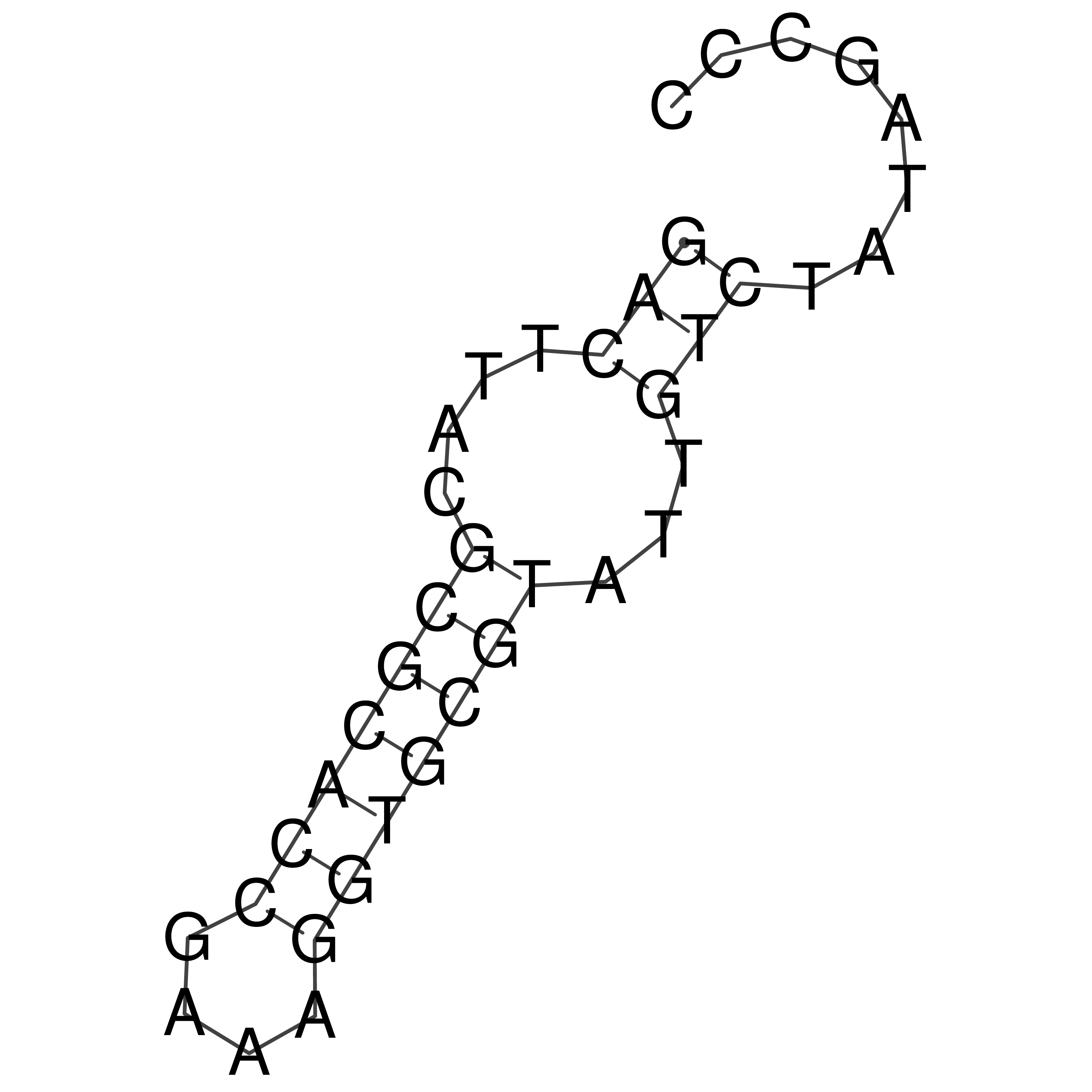
Reference
[1] Revilla C et al. (2008) Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother. 52(4):1472-80. [PMID:18268088]
Relaxase
| ID | 211 | GenBank | YP_001874877 |
| Name | TrwC_R7K |
UniProt ID | B2G2P0 |
| Length | 966 a.a. | PDB ID | |
| Note | conjugative relaxase | ||
Relaxase protein sequence
Download Length: 966 a.a. Molecular weight: 107460.80 Da Isoelectric Point: 9.8804
MLSHMVLTRQDIGRAASYYEDGADDYYAKDGDASEWQGKGAEELGLSGEVDSKRFRELLAGNIGEGHRIM
RSATRQDSKERIGLDLTFSAPKSVSLQALVAGDAEIIKAHDRAVARTLEQAEARAQARQKIQGKTRIETT
GNLVIGKFRHETSRERDPQLHTHAVILNMTKRSDGQWRALKNDEIVKATRYLGAVYNAELAHELQKLGYQ
LRYGKDGNFDLAHIDRQQIEGFSKRTEQIAEWYAARGLDPNSVSLEQKQAAKVLSRAKKTSVDREALRAE
WQATAKELGIDFSRREWSGREKGGSEKQAHSFMPSDEAAKRAVRYAINHLTERQSVMDERELVDTAMKHA
VGAARLEDIQKELLRQTETGYLIREAPRYRPGGQTGPTDEPGKTRAEWIAELAAKGMKQGAARERVDNAI
KTGGLVPIEPRYTTQTALEREKRILQIERDGRGAVAPVIAAEAARERLASTNLNQGQREAAELIVSAANR
VVGVQGFAGTGKSHMLDTAKQMIEGEGYHVRALAPYGSQVKALRELNVEANTLASFLRAKDKNIDSRTVL
VIDEAGVVPTRLMEQTLKLAEKAGARVVLMGDTAQTKAIEAGRPFDQLQAAGMQTAHMREIQRQKNPELK
IAVELAAAGKASSSLERIKDVTEIKNHHERRAAVAEAYIALKPDERDRTLIVSGTNEARREINQIVREGL
GTAGKGIEFDTLVRVDTTQAERRHSKNYQVGHVIQPERDYAKTGLQRGELYRVVETGPGNRLTVIGEHDG
QRIQFSPMTHTKISVYQPERAELAVGDTIRITRNDKHLDLANGDRMKVVAVEDRKVTVTDGKRNVELPTD
KPLHVDHAYATTVHSSQGLTSDRVLIDAHAESRTTAKDVYYVAISRARFEARVFTNDRGKLPAAIARENI
KSAAHDLARDRGGRSAAAERQREQQREAERNRQTQQPAHDRQKAAREAERGMEAGR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B2G2P0 |
Reference
[1] Revilla C et al. (2008) Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother. 52(4):1472-80. [PMID:18268088]
Auxiliary protein
| ID | 297 | GenBank | YP_001874875 |
| Name | TrwA_R7K |
UniProt ID | B2G2N8 |
| Length | 121 a.a. | PDB ID | _ |
| Note | mobilisation protein | ||
Auxiliary protein sequence
Download Length: 121 a.a. Molecular weight: 13473.38 Da Isoelectric Point: 5.0435
MALGEPIKFRLTPEKHAQYEDEAARLGKPLGTYLRERLEADDAVRDELAALRREVVSLHHVIEDLADTGL
RSDQSGPGPNAVQIETLLLLRAIAGPERMKPVKGELKRLGIEVWTPEGKED
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B2G2N8 |
Reference
[1] Revilla C et al. (2008) Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother. 52(4):1472-80. [PMID:18268088]
T4CP
| ID | 211 | GenBank | YP_001874876 |
| Name | TrwB_R7K |
UniProt ID | B2G2N9 |
| Length | 507 a.a. | PDB ID | _ |
| Note | Type IV secretion-system coupling protein | ||
T4CP protein sequence
Download Length: 507 a.a. Molecular weight: 56287.02 Da Isoelectric Point: 9.9420
MHPDDQRKVSAGIVIVLPLIFWITAVQKTEVLGSPKLLALWELMKLTPQKPILLLSALGGLAVGVLFVWL
LNSVGQGEFGGAPFKRFLRGTRIVSGGKLKRMTREKAKQVTVAGVPMPRDAEPRHLLVNGATGTGKSVLL
RELAYTGLLRGDRMVIVDPNGDMLSKFGRDKDVILNPYDKRTKGWSFFNEIRNDYDWQRYALSVVPRGKT
DEAEEWASYGRLLLRETAKKLALIGTPSMRELFHWTTIATFDDLRGFLEGTLAESLFAGSNEASKALTSA
RFVLSDKLPEHVTMPDGDFSIRSWLEDPNGGNLFITWREDMGPALRPLISAWVDVVCTSILSLPEEPKRR
LWLFIDELASLEKLASLADALTKGRKAGLRVVAGLQSTSQLDDVYGVKEAQTLRASFRSLVVLGGSRTDP
KTNEDMSLSLGEHEVERDRYSKNTGKHHSTGRALERVRERVVMPVEIANLPDLTAYVGFAGNRPIAKVPL
EIKQFANRQPAFVEGTI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B2G2N9 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9667..24301
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HXE96_RS00035 | 6438..6773 | + | 336 | WP_181408544 | stable inheritance protein | - |
| HXE96_RS00040 (R7K_007) | 6921..7172 | + | 252 | WP_012196438 | hypothetical protein | - |
| HXE96_RS00045 (R7K_008) | 7327..7713 | - | 387 | WP_012196437 | hypothetical protein | - |
| HXE96_RS00050 (R7K_009) | 7727..8422 | - | 696 | WP_012414157 | StbB family protein | - |
| HXE96_RS00055 (R7K_010) | 8419..8844 | - | 426 | WP_012196435 | hypothetical protein | - |
| HXE96_RS00060 (R7K_011) | 9300..9665 | + | 366 | WP_012414158 | hypothetical protein | - |
| HXE96_RS00065 (R7K_012) | 9667..11190 | + | 1524 | WP_012414159 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| HXE96_RS00070 (R7K_013) | 11202..14090 | + | 2889 | WP_238838742 | MobF family relaxase | - |
| HXE96_RS00075 (R7K_014) | 14173..15249 | - | 1077 | WP_012196431 | P-type DNA transfer ATPase VirB11 | virB11 |
| HXE96_RS00080 (R7K_015) | 15215..16402 | - | 1188 | WP_012414162 | type IV secretion system protein VirB10 | virB10 |
| HXE96_RS00085 (R7K_016) | 16402..17202 | - | 801 | WP_012414163 | P-type conjugative transfer protein VirB9 | virB9 |
| HXE96_RS00090 (R7K_017) | 17214..17909 | - | 696 | WP_012196428 | VirB8/TrbF family protein | virB8 |
| HXE96_RS00220 (R7K_018) | 17914..18048 | - | 135 | Protein_18 | TrwH protein | - |
| HXE96_RS00095 (R7K_019) | 18161..19189 | - | 1029 | WP_012414166 | type IV secretion system protein | virB6 |
| HXE96_RS00100 (R7K_020) | 19203..19433 | - | 231 | WP_012196425 | EexN family lipoprotein | - |
| HXE96_RS00105 (R7K_021) | 19430..20119 | - | 690 | WP_012414167 | P-type DNA transfer protein VirB5 | virB5 |
| HXE96_RS00110 (R7K_022) | 20116..22587 | - | 2472 | WP_012414168 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HXE96_RS00115 (R7K_023) | 22590..22901 | - | 312 | WP_181408538 | VirB3 family type IV secretion system protein | virB3 |
| HXE96_RS00120 (R7K_024) | 22914..23219 | - | 306 | WP_181408539 | VirB2 family type IV secretion system major pilin TrwL | virB2 |
| HXE96_RS00125 (R7K_025) | 23185..23481 | - | 297 | WP_012414171 | KorA family transcriptional regulator | - |
| HXE96_RS00130 (R7K_026) | 23603..24301 | - | 699 | WP_012414172 | lytic transglycosylase domain-containing protein | virB1 |
| HXE96_RS00135 (R7K_027) | 24303..24689 | - | 387 | WP_238838740 | hypothetical protein | - |
| HXE96_RS00140 (R7K_028) | 25033..25320 | + | 288 | WP_012414174 | H-NS histone family protein | - |
| HXE96_RS00145 (R7K_029) | 25356..25769 | + | 414 | WP_012414175 | hypothetical protein | - |
| HXE96_RS00150 (R7K_030) | 25921..26577 | + | 657 | WP_012414176 | recombinase family protein | - |
| HXE96_RS00155 (R7K_031) | 26570..27541 | + | 972 | WP_012414177 | replication protein RepA | - |
| HXE96_RS00160 (R7K_032) | 28255..28884 | + | 630 | WP_181408540 | DNA-binding protein | - |
Host bacterium
| ID | 211 | GenBank | NC_010643 |
| Plasmid name | R7K | Incompatibility group | IncW |
| Plasmid size | 39792 bp | Coordinate of oriT [Strand] | 9136..9174 [-] |
| Host baterium | Providencia rettgeri |
Cargo genes
| Drug resistance gene | blaTEM-1D, aph(6)-Id, aph(3'')-Ib, aadA13 |
| Virulence gene | trwD, trwF, trwG, trwK, trwM |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |