Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100197 |
| Name | oriT_pMRC01 |
| Organism | Lactococcus lactis |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_001949 (1811..1848 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..11, 14..25 (ACACCACCCAA..TTGGAGTGGTGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
TAAGTGCGCATT |
| Note | The oriT region was determined experimentally, but the putative nic site was not determined experimentally. |
oriT sequence
Download Length: 38 nt
ACACCACCCAATTTTGGAGTGGTGTGTAAGTGCGCATT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file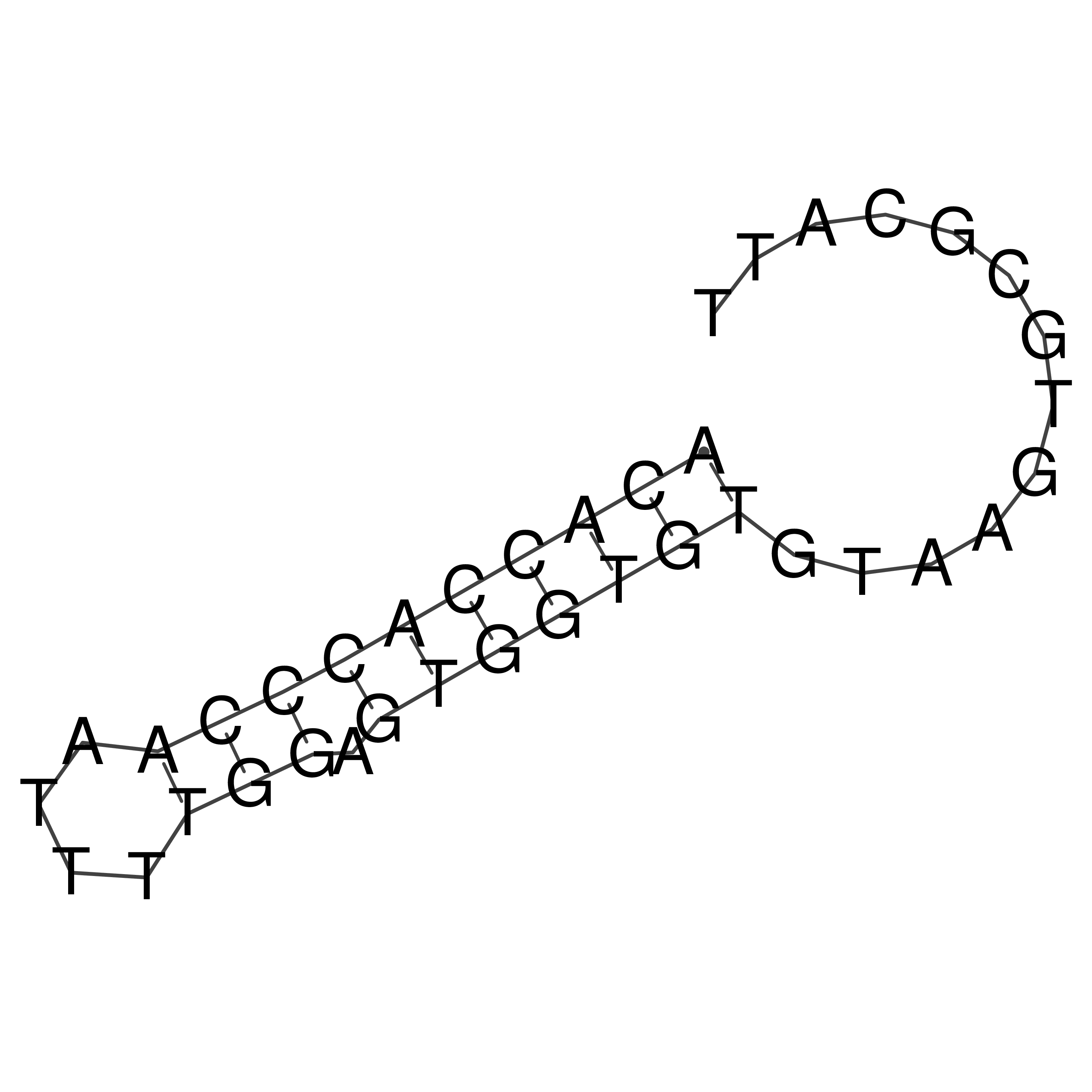
Reference
[1] Parker C et al. (2005) Elements in the co-evolution of relaxases and their origins of transfer. Plasmid . 53(2):113-8. [PMID:15737398]
[2] Becker EC et al. (2003) Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J Bacteriol. 185(12):3538-46. [PMID:12775691]
[3] Hickey RM et al. (2001) Exploitation of plasmid pMRC01 to direct transfer of mobilizable plasmids into commercial lactococcal starter strains. Appl Environ Microbiol. 67(6):2853-8. [PMID:11375207]
[4] Dougherty BA et al. (1998) Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 29(4):1029-38. [PMID:9767571]
Relaxase
| ID | 190 | GenBank | NP_047290 |
| Name | TraA_pMRC01 |
UniProt ID | O87207 |
| Length | 680 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 680 a.a. Molecular weight: 79275.45 Da Isoelectric Point: 6.7052
MAIFHMNFSNISAGKGRSAVASASYRSGEKLYSEMENKTYFYNRSVMPESFILLPENAPEWAKDRQKLWN
EVEAVDRKVNSRYAKEFNVALPIELSEDEQKELLTEYVQKIFVDKGMVADVAIHRDHDENPHAHVMLTNR
PFNADGSWGQKAKKEYILDENGNKTYTANGHARSRKIWLVDWDKVGKVEEWRKAWADHVNSVFQEKNIDE
RISEKTLEAQGINDIATQHVGVTGNRDERAEFNKLVLENRQHKAELENLDEKINNELKVKQLKNFYSFNE
KKVIAELSKELHTFIDLEHLEEKNKMLFNWKNSVLIKQIVGKDVSEELNKVSRQEFSLDNANRLIDKVVD
RSIKAMYPTVDSNAFSMPEKRQLIRETESENKVFTGAELDDRLSMIRADSLNRQIVALTKRPFTSLTMLS
KQEKYHIDNINNVLAYQGYNYENIEMSHGKILRNYESEEFSKELDIIQRSLKQINSIEEVRKIVEQQYSV
VLGTIFPDSELNKLSLVEKEKTYNAVIYFNPTIKKLTQSELENIVKEPPILFSTKEHNQGLSYLAGRIRL
EDINNKHLTRVLKFEGTKKLFIGEAKADKNIDPELLKEATNRNDKQSYKNEFYRDKNMENYQSVAYSDTS
PAEYMNRLFSGVLISVLYNNENQHKKGLEEVEWDMERKHREHTKTGGLSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | O87207 |
Reference
[1] Parker C et al. (2005) Elements in the co-evolution of relaxases and their origins of transfer. Plasmid . 53(2):113-8. [PMID:15737398]
[2] Hickey RM et al. (2001) Exploitation of plasmid pMRC01 to direct transfer of mobilizable plasmids into commercial lactococcal starter strains. Appl Environ Microbiol. 67(6):2853-8. [PMID:11375207]
[3] Dougherty BA et al. (1998) Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 29(4):1029-38. [PMID:9767571]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5386..15263
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS157_RS00010 (pMRC01_002) | 1099..1365 | - | 267 | WP_010890660 | hypothetical protein | - |
| HS157_RS00015 (pMRC01_004) | 1378..1662 | - | 285 | WP_010890661 | hypothetical protein | - |
| HS157_RS00020 (pMRC01_005) | 1931..3973 | + | 2043 | WP_010890626 | MobQ family relaxase | - |
| HS157_RS00025 (pMRC01_006) | 4058..4402 | + | 345 | WP_010890662 | hypothetical protein | - |
| HS157_RS00030 (pMRC01_007) | 4403..5008 | + | 606 | WP_010890663 | hypothetical protein | - |
| HS157_RS00035 (pMRC01_008) | 5021..5365 | + | 345 | WP_010890627 | CagC family type IV secretion system protein | - |
| HS157_RS00040 (pMRC01_009) | 5386..5733 | + | 348 | WP_010890628 | hypothetical protein | trsC |
| HS157_RS00045 (pMRC01_010) | 5717..6370 | + | 654 | WP_010890629 | TrsD/TraD family conjugative transfer protein | trsD |
| HS157_RS00050 (pMRC01_011) | 6382..8400 | + | 2019 | WP_010890630 | DUF87 domain-containing protein | virb4 |
| HS157_RS00055 (pMRC01_012) | 8393..9811 | + | 1419 | WP_010890631 | type VII secretion protein EssB/YukC | - |
| HS157_RS00060 (pMRC01_013) | 9813..10970 | + | 1158 | WP_010890632 | phage tail tip lysozyme | prgK |
| HS157_RS00065 (pMRC01_014) | 10983..11600 | + | 618 | WP_010890664 | hypothetical protein | - |
| HS157_RS00070 (pMRC01_015) | 11590..11958 | + | 369 | WP_010890633 | hypothetical protein | - |
| HS157_RS00075 (pMRC01_016) | 11955..12413 | + | 459 | WP_010890634 | hypothetical protein | trsJ |
| HS157_RS00080 (pMRC01_017) | 12410..14002 | + | 1593 | WP_010890635 | VirD4-like conjugal transfer protein, CD1115 family | - |
| HS157_RS00085 (pMRC01_018) | 14017..14412 | + | 396 | WP_010890665 | hypothetical protein | - |
| HS157_RS00090 (pMRC01_019) | 14427..15263 | + | 837 | WP_010890636 | conjugal transfer protein TrbL family protein | trsL |
| HS157_RS00095 (pMRC01_020) | 15279..17456 | + | 2178 | WP_010890637 | type IA DNA topoisomerase | - |
| HS157_RS00100 (pMRC01_021) | 17459..17668 | + | 210 | WP_010890684 | hypothetical protein | - |
| HS157_RS00105 (pMRC01_022) | 17671..18738 | + | 1068 | WP_010890638 | ArdC family protein | - |
| HS157_RS00110 (pMRC01_023) | 18786..19238 | - | 453 | WP_010890666 | hypothetical protein | - |
| HS157_RS00115 (pMRC01_024) | 19521..19721 | - | 201 | WP_010890667 | hypothetical protein | - |
Host bacterium
| ID | 190 | GenBank | NC_001949 |
| Plasmid name | pMRC01 | Incompatibility group | IncQ-type |
| Plasmid size | 60232 bp | Coordinate of oriT [Strand] | 1811..1848 [-] |
| Host baterium | Lactococcus lactis |
Cargo genes
| Drug resistance gene | _ |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |