Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100195 |
| Name | oriT_pIJB1 |
| Organism | Burkholderia cepacia |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_019378 (_) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 13..31, 34..52 (GTGAAGATAGATAACCGGC..GCCGGTTAGCTAACTTCAC) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
CATCCTG|C |
| Note | _ |
oriT sequence
Download Length: 99 nt
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file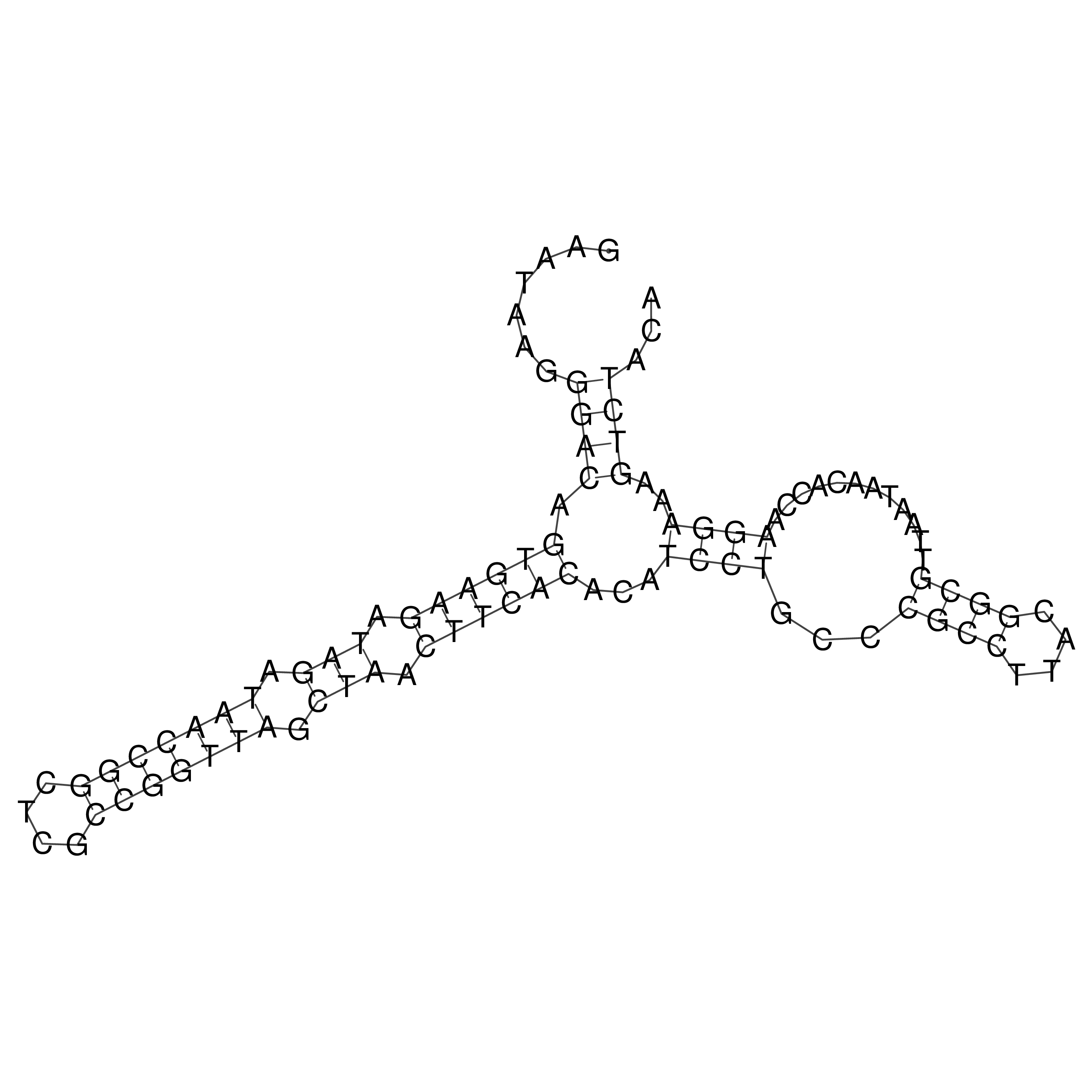
Relaxase
| ID | 188 | GenBank | YP_006965894 |
| Name | TraI_pIJB1 |
UniProt ID | K4IYE5 |
| Length | 730 a.a. | PDB ID | |
| Note | TraI Conjugative transfer protein, DNA relaxase | ||
Relaxase protein sequence
Download Length: 730 a.a. Molecular weight: 80015.48 Da Isoelectric Point: 10.3609
MIAKHVSMKTVQKSDFAGLVKYITDEQAKNERVGYVAVTNCHTDRPEVAITEVLNTQAQNTRSGADKTYH
LIVSFRPGEQPDDATLKAIEARICEGLGFGEHQRVSAVHHDTDNLHIHIAINKIHPTRYTILEPFNAYHT
LGQLCEKLEREYGLERDNHQGNKLVSEGRAQDMERHAGVESLQGWVKRECAADMQAAQSWEALHQVMRDN
GLELRERGNGFVIQAADGLAVKASSIGREFSKAKLEERLGTFEPSAEHLERAAQKPRKQYDKKPVRSRVN
TVELHAKYKADQLTISGSRTAEWEKARAKKDRLIEAAKRSGRLKRSTIKLISGAGVGKKLMYSATSKTMK
SEIEKINKQYLKERQEIHEKYQRRAWADWLRAKAAEGDQEALGALRARDAAQGLKGNTVAGTGPLRPQAA
AAEQDSVTKKGTIIYRVGPTAVRDDGDRLKVSRGADQDGMQAALRMAMDRYGDRISVGGTPAFKEQIAQA
AAASKLPITFDDDALERRRQELLTQATTKENDHDRTDRGRAAGGGISGSGPAIAAIPGAARAGQPRAAAA
GRTTKPNVGRVGRKPPPQSQNRLRRLSQLGVVQLASGSEVLLPRHVPGDVEQQGAKPVDGLRRDIFGPGR
LSPQVAAAADKYIAEREGKRLKGFDIPKHSRYTDGQDGAATFAGIRQVEGQSLALLTRGEEVMVLPVDEP
TARRLKRLAVGDAITVTPQGSIKTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K4IYE5 |
Auxiliary protein
| ID | 266 | GenBank | YP_006965895 |
| Name | TraH_pIJB1 |
UniProt ID | K4J0Y9 |
| Length | 108 a.a. | PDB ID | _ |
| Note | TraH Conjugative transfer protein, relaxosome stabilization | ||
Auxiliary protein sequence
Download Length: 108 a.a. Molecular weight: 11836.18 Da Isoelectric Point: 3.8732
MTEPTEDELLEAALAAVDQPLPQSPEPPEPDSHEPPQPDGPPSPTLAALDESRRPKAKTVCEDCPNSVWF
SSPAEVKCYCRVMFLVTWSNKEPNQLTACDGIFLGQED
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K4J0Y9 |
| ID | 267 | GenBank | YP_006965896 |
| Name | TraJ_pIJB1 |
UniProt ID | Q2VLB1 |
| Length | 129 a.a. | PDB ID | _ |
| Note | TraJ Conjugative transfer protein, relaxosome protein | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14479.81 Da Isoelectric Point: 10.3612
MAKTRNSDDPRMPTRKAGRHLRVPVLPDEEAEIKRLAASVGLPVAAYLRNVGLGYQVPGILDNKRVEELA
RINGDLGRLGGLLKLWLTDDVRTLQFGETTILALLSRIESTQDRMHEVMQEVVTPRVKR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q2VLB1 |
| ID | 268 | GenBank | YP_006965897 |
| Name | TraK_pIJB1 |
UniProt ID | K4JDT7 |
| Length | 132 a.a. | PDB ID | _ |
| Note | TraK conjugative transfer protein,oriT binding protein | ||
Auxiliary protein sequence
Download Length: 132 a.a. Molecular weight: 14842.04 Da Isoelectric Point: 10.5392
MPKSYPEELAEWVKKREATRPRQDKNVVAFLALKSDVQDAIAAGYSLRTIWEHMHEKGKIPYRYETFLKH
VRRHIKGRPPAVVDGQPSTPPDPPRPPAAGKTAAPRPQQPTKSAPAPAGFKFDSQPKKEDLL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | K4JDT7 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1821..12421
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTB43_RS00005 (D704_p36) | 1..870 | - | 870 | WP_015063508 | plasmid replication initiator TrfA | - |
| HTB43_RS00010 (D704_p34) | 1191..1568 | + | 378 | WP_011171717 | transcriptional regulator | - |
| HTB43_RS00015 (D704_p33) | 1821..2783 | + | 963 | WP_011171718 | P-type conjugative transfer ATPase TrbB | virB11 |
| HTB43_RS00020 (D704_p32) | 2795..3223 | + | 429 | WP_011171719 | IncP-type conjugal transfer pilin TrbC | virB2 |
| HTB43_RS00025 (D704_p31) | 3226..3537 | + | 312 | WP_011171720 | conjugal transfer protein TrbD | virB3 |
| HTB43_RS00030 (D704_p30) | 3534..6071 | + | 2538 | WP_015063520 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HTB43_RS00035 (D704_p29) | 6068..6844 | + | 777 | WP_015063521 | conjugal transfer protein TrbF | virB8 |
| HTB43_RS00040 (D704_p28) | 6857..7762 | + | 906 | WP_015063522 | P-type conjugative transfer protein TrbG | virB9 |
| HTB43_RS00045 (D704_p27) | 7765..8232 | + | 468 | WP_015063523 | TrbH Mating pair formation protein | - |
| HTB43_RS00050 (D704_p26) | 8238..9632 | + | 1395 | WP_015063524 | TrbI/VirB10 family protein | virB10 |
| HTB43_RS00055 (D704_p25) | 9651..10424 | + | 774 | WP_015063525 | P-type conjugative transfer protein TrbJ | virB5 |
| HTB43_RS00060 (D704_p24) | 10435..10674 | + | 240 | WP_015063526 | entry exclusion lipoprotein TrbK | - |
| HTB43_RS00065 (D704_p35) | 10685..12421 | + | 1737 | WP_172685318 | P-type conjugative transfer protein TrbL | virB6 |
| HTB43_RS00070 (D704_p23) | 12441..13034 | + | 594 | WP_015063527 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HTB43_RS00075 (D704_p22) | 13048..13653 | + | 606 | WP_015063528 | transglycosylase SLT domain-containing protein | - |
| HTB43_RS00080 (D704_p02) | 13679..13942 | + | 264 | WP_015063529 | hypothetical protein | - |
| HTB43_RS00085 (D704_p01) | 13945..14643 | + | 699 | WP_015063530 | TraX family protein | - |
| HTB43_RS00090 | 14644..15000 | + | 357 | WP_258404801 | OmpA family protein | - |
| HTB43_RS00095 (D704_p55) | 15086..15571 | + | 486 | WP_015063510 | thermonuclease family protein | - |
| HTB43_RS00100 (D704_p56) | 15556..15975 | + | 420 | WP_172685320 | hypothetical protein | - |
| HTB43_RS00105 (D704_p54) | 15932..16303 | - | 372 | WP_015063512 | resolvase | - |
Host bacterium
| ID | 188 | GenBank | NC_019378 |
| Plasmid name | pIJB1 | Incompatibility group | IncP-1 |
| Plasmid size | 99001 bp | Coordinate of oriT [Strand] | 43131..43230 [+] |
| Host baterium | Burkholderia cepacia |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR1, merT, merP, merA, merB, merR2, merD, merE |
| Degradation gene | linF |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9, AcrIF24 |