Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100192 |
| Name | oriT_pUB110 |
| Organism | Staphylococcus aureus |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_001384 (1144..1181 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 2..8, 13..19 (ACTTTAT..ATAAAGT) |
| Location of nic site | 27..28 |
| Conserved sequence flanking the nic site |
TAGTGTG|TTAT |
| Note | _ |
oriT sequence
Download Length: 38 nt
>oriT_pUB110
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTAC
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file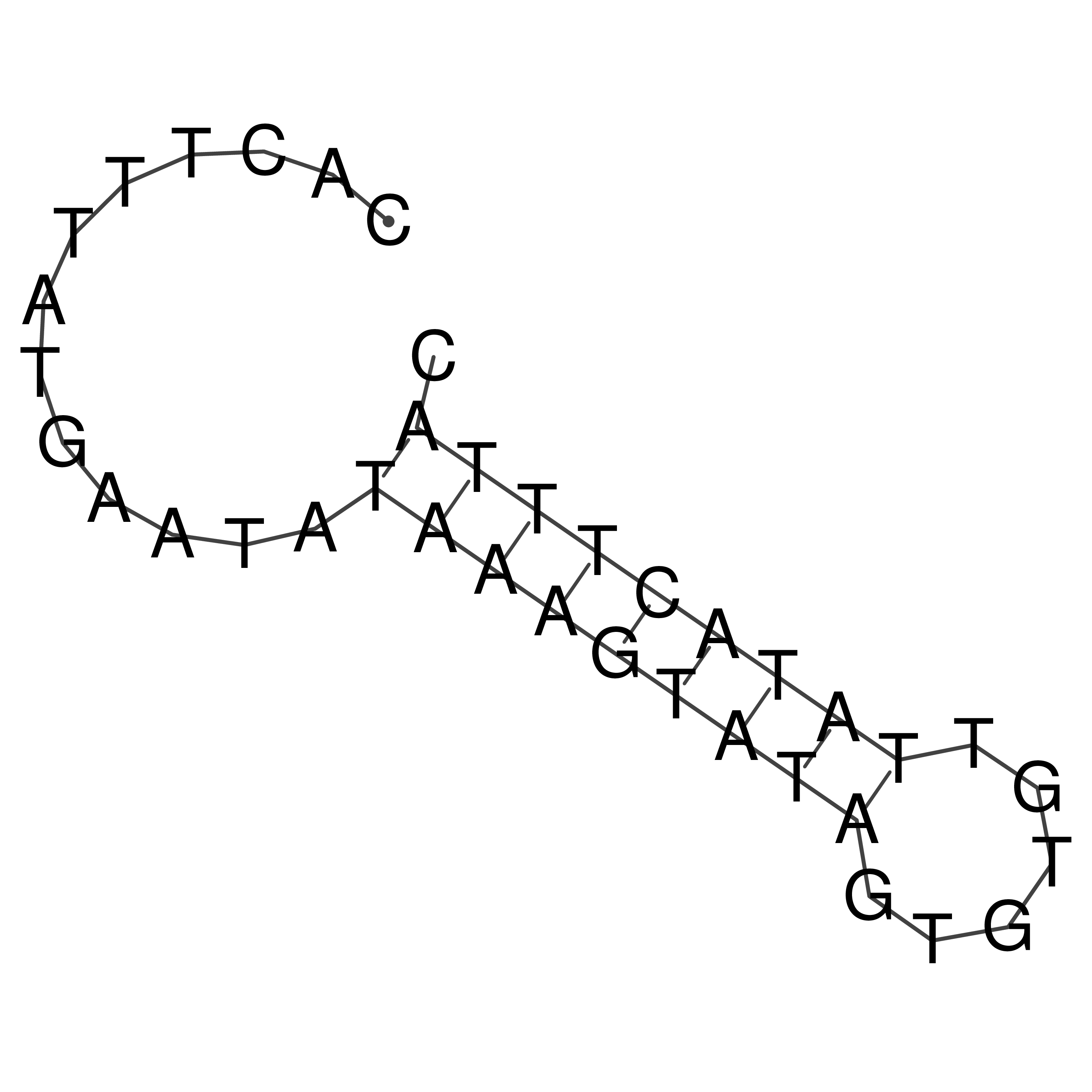
Reference
[1] Grohmann E et al. (2003) Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 67(2):277-301. [PMID:12794193]
Relaxase
| ID | 185 | GenBank | NP_040431 |
| Name | Mob_pUB110 |
UniProt ID | Q9KJC8 |
| Length | 420 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 420 a.a. Molecular weight: 49660.33 Da Isoelectric Point: 9.9693
>NP_040431.1 beta protein (plasmid) [Staphylococcus aureus]
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9KJC8 |
Host bacterium
| ID | 185 | GenBank | NC_001384 |
| Plasmid name | pUB110 | Incompatibility group | - |
| Plasmid size | 4548 bp | Coordinate of oriT [Strand] | 1144..1181 [-] |
| Host baterium | Staphylococcus aureus |
Cargo genes
| Drug resistance gene | aadD |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |