Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100182 |
| Name | oriT_pTC-F14 |
| Organism | Acidithiobacillus caldus |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_004734 (2024..2073 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 3..18, 22..37 (CTTGGCGGGAACATGC..GCATGTGTATGACAAG) |
| Location of nic site | 47..48 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | _ |
oriT sequence
Download Length: 50 nt
GGCTTGGCGGGAACATGCGTAGCATGTGTATGACAAGTCCCATCCTGTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file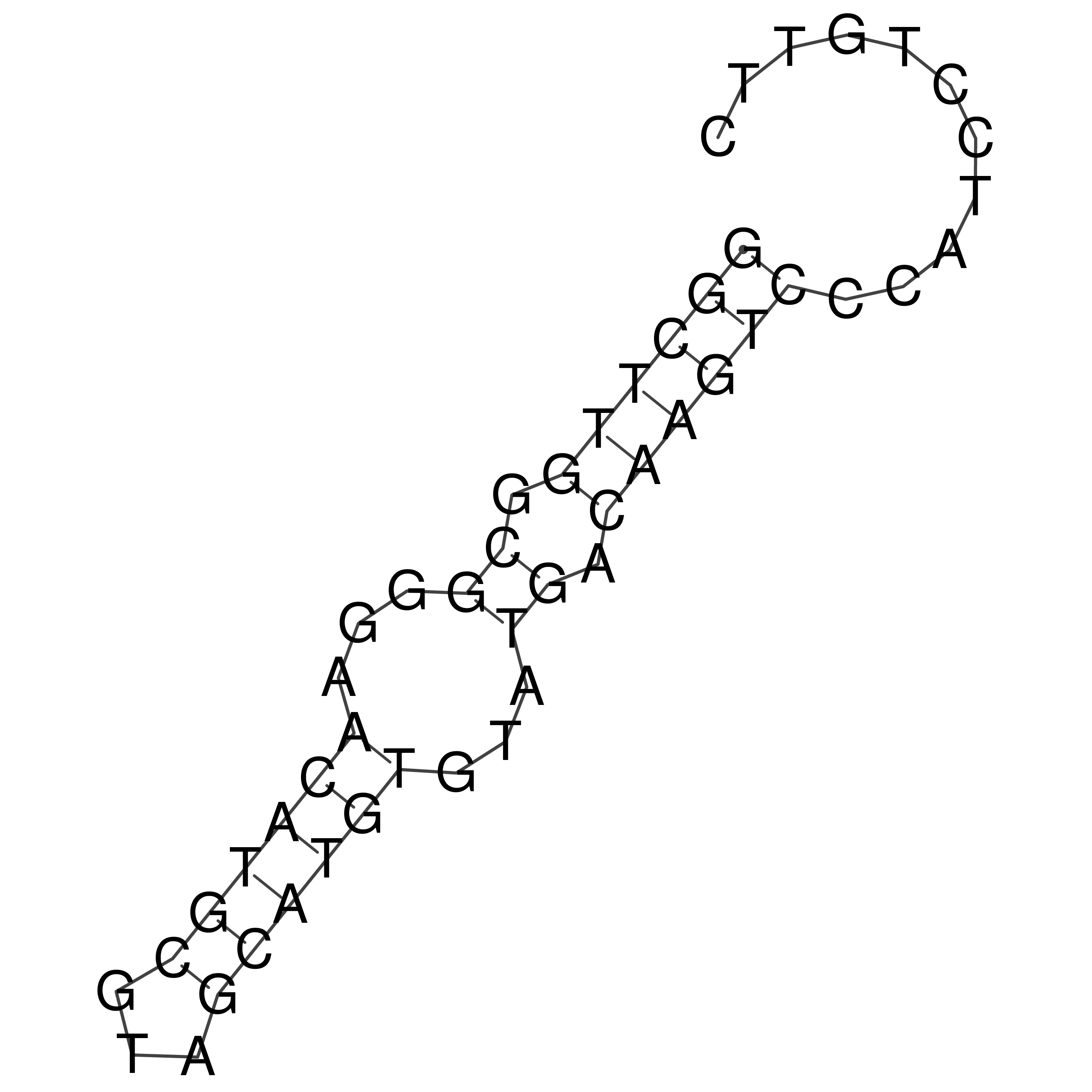
Reference
[1] Rawlings DE (2005) The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid. 53(2):137-47. [PMID:15737401]
[2] van Zyl LJ et al. (2003) Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J Bacteriol. 185(20):6104-11. [PMID:14526022]
Relaxase
| ID | 175 | GenBank | NP_835376 |
| Name | MobA_pTC-F14 |
UniProt ID | Q840R3 |
| Length | 889 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 889 a.a. Molecular weight: 102296.78 Da Isoelectric Point: 9.8238
MIVKKVKSNRTKGKAASIRDLTNYIREPQNRNPNEKVLYANGRGFISDTHAAQREEMVALAAEAVRSRNP
VNHYILSWREGEQPSPEQVEEAVSIFLDELGLQEHQVIYALHKDTDNLHLHIAVNRVHPETLKCVEINKG
FDLESAHRAIARIEHAQGWQREQNGRYEVLENGELGREHLEPNKRRQPEQRKRDKENRTGEKSAERIAIE
IGAPIIKQAQSWEQLHRELAAQGMRYEQKGSGALLWVGEVAVKASSADREASLGKLQKRLGAYEPAQAPS
PVAQRKPEPLQPDRPEWEDFMAGRKMHYAEKNAAKLSMDQRQEQERKALQARQQEQRKVLLGGRWNGKGE
ALNALRGVLAAEQAAEKAALKERHQQERQQWRQQYRPYPDFEQWLRREHGAEQATRWRYREAEPQRIEGD
STEAPKPRDIRAYRAEIVGQEVRYTPKSGAGGGPGGVSFVDKGRIIEIHDWRNQDTTLAALQLSAQKWGK
FTVTGNDEYKALCVKLAVEHGFQITNPELQEVIRQERQRMRQERAQAMKSEQIKPFERYAEAVGAERYRV
TSIKMRPDGSKQTFILDKRDGITRGFTPEEIAQRTPEMQRLQRRGENLYYTPLSEGKHHILIDDMDREKL
DRLIRDGYQPAVVLESSPGNYQAVITIPKLGTPFDKDVGNRLSDALNREYGDPKLSGAIHPHRAPGYENR
KPKHRREDGSYPEVRLLKAERRECGKTLALSREIDAGYQRQAAEKALKTPVEQFSEPNSTQTTPVSEKTA
TEAYWRHYRDVRKRQRGMLDLSRVDAMIAVRMRVTGFDQSAIEGAIYQCAPSIREQQESRDWTDYARRTA
RYAYSAAGDRQAADLGKYRQQWEKLEGRERQQEQAKAREIERNGPSMSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q840R3 |
Reference
[1] Rawlings DE (2005) The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid. 53(2):137-47. [PMID:15737401]
[2] van Zyl LJ et al. (2003) Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J Bacteriol. 185(20):6104-11. [PMID:14526022]
Auxiliary protein
| ID | 252 | GenBank | NP_835375 |
| Name | MobB_pTC-F14 |
UniProt ID | Q840R4 |
| Length | 103 a.a. | PDB ID | _ |
| Note | oriT recognition-like protein | ||
Auxiliary protein sequence
Download Length: 103 a.a. Molecular weight: 11210.91 Da Isoelectric Point: 9.7756
MPFTVQGLEPLDAVVNVRLTASEKARLREDADLAGLSVSELVRRRYFGRPIVAHADAVLLKELRRIGGLL
KHVHNESGGAYSQQTAAVLVTLKAAIEGLSHDR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q840R4 |
Reference
[1] Rawlings DE (2005) The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid. 53(2):137-47. [PMID:15737401]
[2] van Zyl LJ et al. (2003) Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J Bacteriol. 185(20):6104-11. [PMID:14526022]
| ID | 253 | GenBank | NP_835374 |
| Name | MobC_pTC-F14 |
UniProt ID | Q840R5 |
| Length | 131 a.a. | PDB ID | _ |
| Note | histone-like mobilization protein | ||
Auxiliary protein sequence
Download Length: 131 a.a. Molecular weight: 13984.96 Da Isoelectric Point: 10.2555
MASSKQLYDIDAYAKKARTALENLEKTQQPGEVTGKGGKADVIRAIKSDLEALIKKGYTSKQIADALKHD
DVFGILPKTITEIVAGKKQNKSTVTRKKKSTTPAPTASGQANTAPIPQAGTFTVKPDSEDL
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q840R5 |
Reference
[1] Rawlings DE (2005) The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid. 53(2):137-47. [PMID:15737401]
[2] van Zyl LJ et al. (2003) Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J Bacteriol. 185(20):6104-11. [PMID:14526022]
Host bacterium
| ID | 175 | GenBank | NC_004734 |
| Plasmid name | pTC-F14 | Incompatibility group | IncQ2 |
| Plasmid size | 14155 bp | Coordinate of oriT [Strand] | 2024..2073 [-] |
| Host baterium | Acidithiobacillus caldus |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |