Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100177 |
| Name | oriT_R721 |
| Organism | Escherichia coli |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_002525 (29007..29059 [-], 53 nt) |
| oriT length | 53 nt |
| IRs (inverted repeats) | 5..21, 25..42 (CGATTGTAACATGACCG..CGGTCTTGTGTACAATCG) |
| Location of nic site | 50..51 |
| Conserved sequence flanking the nic site |
TATCGTG|C |
| Note | _ |
oriT sequence
Download Length: 53 nt
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file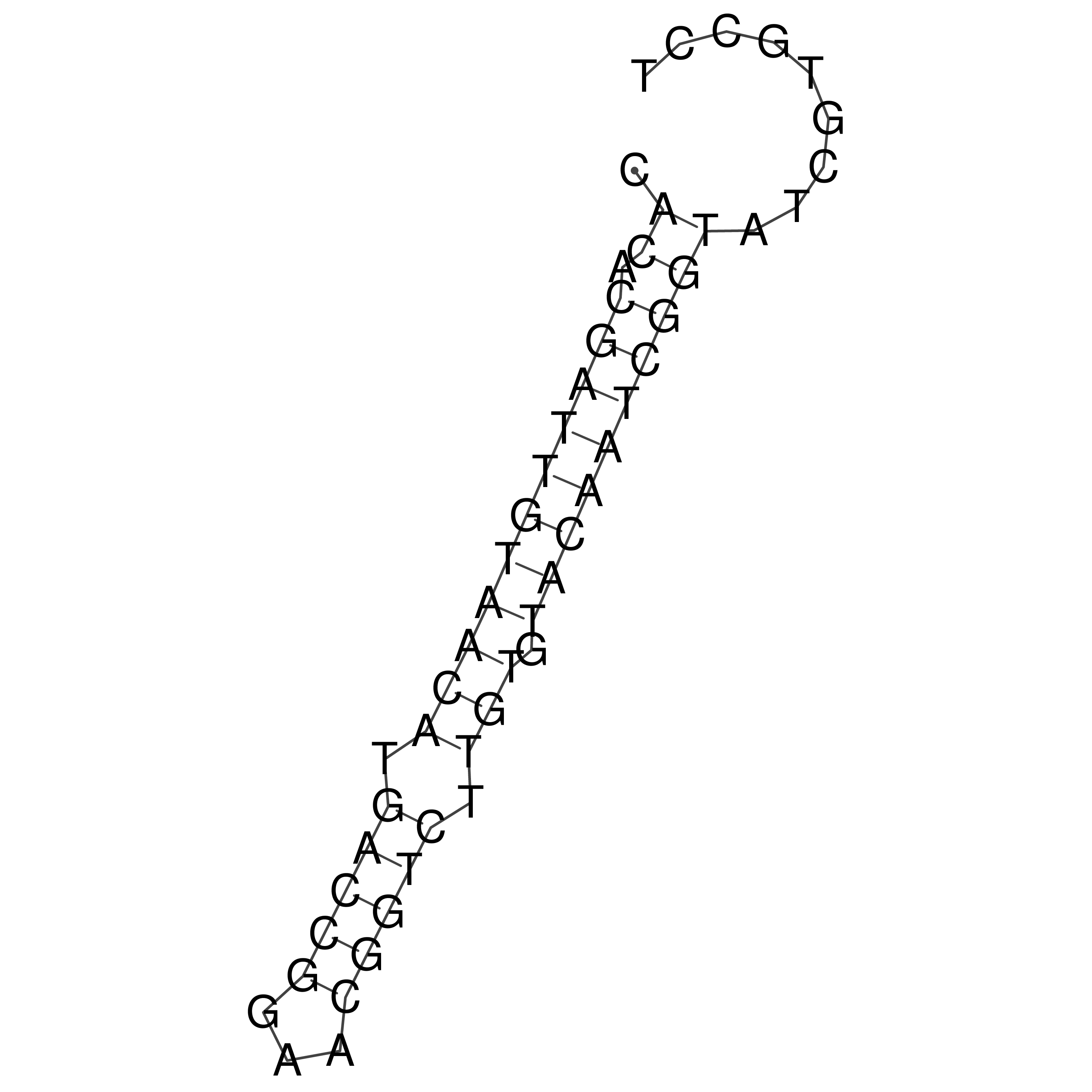
Reference
[1] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[2] Kim SR et al. (1992) Nucleotide sequence of the R721 shufflon. J Bacteriol. 174(21):7053-8. [PMID:1400257]
Relaxase
| ID | 170 | GenBank | NP_065331 |
| Name | NikB_R721 |
UniProt ID | Q9F553 |
| Length | 1272 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 1272 a.a. Molecular weight: 145804.44 Da Isoelectric Point: 5.1637
MIVRYGGGNDGIVDYLINGRKAERQYTRDELDHRVVLDGDLQTTDKIIDSIENKSQERYLHITLSFHESH
VSNEVLKAVVDDYKKLLMNAYHPDEYSFYAEAHLPKIRHIQDNSTGELVERKPHIHIVIPKVNLITEKFL
NPVGDVTKGHTIEQLDAIQEFINNKYNLDSPKDYPRKDADYGKIISRVKGDLYKEHHSELKGELLSRIEN
EKIENYSVFKDIVAEYGELRIRNAGKTNEYLAVKLPGDKKYTNLKSPLFRQNYIETRTLTLEKPTHKEIE
KRLNAWLNKTSHEIKHIFNQAEKTRELYKTLSPSQQIDFLQERIKEYDSREKLNERNSQQTSGRAGGYKS
CPKKFARIRQSEATVGLSRMPQRGMVYGINGFTRPDSVSVLSDISQRDLAKQLPQREHPGQDVRRDYDRQ
FTESGIKSLERSSFLCETMFQTLNEAAEKNEITTMAEIRRNIDPVRFLSSAAERFNIIPAQHKIRTAKDG
SPRFSVGNRNMNASDFLTKHINLAWKDAKSFLLEVYSQQLENTPYTRYPTYRRLTHHEARERLNSLNLSE
KTLRNTIRFERGKLYNDLREMRRELKLIPREQRDIAVGVIVYKKLTTLERLSELDTEGRHIIRQYHADWH
KDKDEMKALERLKSYLNFDEINAISADEPELSLQKAVDSQRRLEEAKKINTKLKDLVMDKQDSRIVYRDQ
ESEKPVFTDKGNFVVAGKNPSKEEIGIMLEYSREKFGGVLKLTGSEDFKKMCAEVAAEQDMKIILRPEQY
QQMMLELKAELQGNKFEQVETQENSQESESRIEKGDALKEQATEQEQATEQAQATEQEQATEQAQATEQA
QATEQAQATEQAQATEQAQATEQAQDTEQAQATEQAQATEQAQATEQAQIAEQATSSYDPGVITRANILD
SQMLSKGTNGEFGYLKSLDSDENEIWEVLGHVPGDSDDIFSVASFDNENDAKEFCKIVNELGIDRTQALI
QEQLSSQQEQAAVQETYQESPEEILASISANEHMISGLENFLVKDRSQFSSCNGDIVVEAEITRGEGGLY
HLAVAGKHGLERGDAVARVDVTEQQFAAITGKTPSEVLTGDQTSARVPVITGIHFSTRAIENVNKLEQQK
DYVYFSTHEGLNAEIKDFSSLKDAIEWGRVECELHDLNKRDTVIYRVESEHISQGIDAVMKNAERVERHE
IEKAQGRDCTPEDGKILEAIDRFEDKFRGEGLKFEREKAESDLLNHGFTREMAEDALGKQFVQAREDHQE
SQQQRDDSQDMH
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9F553 |
Auxiliary protein
| ID | 245 | GenBank | NP_065330 |
| Name | NikA_R721 |
UniProt ID | Q9F554 |
| Length | 113 a.a. | PDB ID | _ |
| Note | relaxosome component | ||
Auxiliary protein sequence
Download Length: 113 a.a. Molecular weight: 13147.21 Da Isoelectric Point: 10.4689
MKKKTNKNVHVTFRLTEEEYAPFDRAIKELNISKSEFFRLLTIGKINTYASDKRNIPEYKRCLSQLSWAG
NNINQIAHRLNSDHLKGIISESLYKKVLNGLIGIRDRLQEIAK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9F554 |
Reference
[1] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46638..70260
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HXE98_RS00265 (R721_54) | 42127..43251 | - | 1125 | WP_000486716 | site-specific integrase | - |
| HXE98_RS00495 | 43664..43885 | - | 222 | Protein_54 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HXE98_RS00275 | 43861..44190 | + | 330 | WP_001141249 | hypothetical protein | - |
| HXE98_RS00280 | 44733..45986 | - | 1254 | WP_010895887 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HXE98_RS00285 (R721_56) | 45999..46634 | - | 636 | WP_000934978 | A24 family peptidase | - |
| HXE98_RS00290 (R721_57) | 46638..47066 | - | 429 | WP_001326801 | lytic transglycosylase domain-containing protein | virB1 |
| HXE98_RS00295 (R721_58) | 47186..47743 | - | 558 | WP_000095048 | type 4 pilus major pilin | - |
| HXE98_RS00300 (R721_59) | 47788..48897 | - | 1110 | WP_000974903 | type II secretion system F family protein | - |
| HXE98_RS00305 (R721_60) | 48888..50441 | - | 1554 | WP_000466228 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HXE98_RS00310 (R721_61) | 50466..50960 | - | 495 | WP_000912555 | type IV pilus biogenesis protein PilP | - |
| HXE98_RS00315 (R721_62) | 50944..52266 | - | 1323 | WP_010895889 | type 4b pilus protein PilO2 | - |
| HXE98_RS00320 (R721_63) | 52305..53948 | - | 1644 | WP_001035589 | PilN family type IVB pilus formation outer membrane protein | - |
| HXE98_RS00325 (R721_64) | 53941..54459 | - | 519 | WP_010895890 | sigma 54-interacting transcriptional regulator | virb4 |
| HXE98_RS00330 (R721_65) | 54506..56464 | - | 1959 | WP_010895891 | type IV secretory system conjugative DNA transfer family protein | - |
| HXE98_RS00335 (R721_66) | 56480..57535 | - | 1056 | WP_001059977 | P-type DNA transfer ATPase VirB11 | virB11 |
| HXE98_RS00340 (R721_67) | 57714..58853 | - | 1140 | WP_000790641 | TrbI/VirB10 family protein | virB10 |
| HXE98_RS00345 (R721_68) | 58843..59574 | - | 732 | WP_044083601 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| HXE98_RS00350 (R721_69) | 59610..60344 | - | 735 | WP_000432283 | type IV secretion system protein | virB8 |
| HXE98_RS00355 (R721_71) | 60510..62867 | - | 2358 | WP_010895892 | VirB4 family type IV secretion system protein | virb4 |
| HXE98_RS00360 (R721_72) | 62873..63193 | - | 321 | WP_010895893 | VirB3 family type IV secretion system protein | virB3 |
| HXE98_RS00365 (R721_73) | 63264..63554 | - | 291 | WP_000865478 | TrbC/VirB2 family protein | virB2 |
| HXE98_RS00370 (R721_74) | 63554..64138 | - | 585 | WP_001177114 | lytic transglycosylase domain-containing protein | virB1 |
| HXE98_RS00375 (R721_75) | 64159..64557 | - | 399 | WP_001153669 | hypothetical protein | - |
| HXE98_RS00380 (R721_76) | 64676..65113 | - | 438 | WP_000539665 | type IV pilus biogenesis protein PilM | - |
| HXE98_RS00385 (R721_77) | 65119..66354 | - | 1236 | WP_000733391 | toxin co-regulated pilus biosynthesis Q family protein | - |
| HXE98_RS00390 (R721_78) | 66357..66644 | - | 288 | WP_029785630 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HXE98_RS00395 (R721_79) | 66816..67451 | - | 636 | WP_000835769 | hypothetical protein | - |
| HXE98_RS00400 (R721_80) | 67499..68308 | + | 810 | WP_001005153 | DUF5710 domain-containing protein | - |
| HXE98_RS00405 (R721_81) | 68361..68615 | - | 255 | WP_000609210 | EexN family lipoprotein | - |
| HXE98_RS00410 (R721_82) | 68618..69259 | - | 642 | WP_001434037 | type IV secretion system protein | - |
| HXE98_RS00415 (R721_83) | 69265..70260 | - | 996 | WP_000276068 | type IV secretion system protein | virB6 |
| HXE98_RS00425 (R721_85) | 70481..70870 | - | 390 | WP_010895894 | hypothetical protein | - |
| HXE98_RS00430 (R721_86) | 71033..71626 | - | 594 | WP_001243169 | hypothetical protein | - |
| HXE98_RS00435 (R721_87) | 71777..72439 | - | 663 | WP_001243159 | hypothetical protein | - |
| HXE98_RS00440 | 72450..72620 | - | 171 | WP_000550721 | hypothetical protein | - |
| HXE98_RS00445 (R721_88) | 72624..73067 | - | 444 | WP_000498521 | NfeD family protein | - |
| HXE98_RS00450 (R721_89) | 73129..74082 | - | 954 | WP_072097371 | SPFH domain-containing protein | - |
| HXE98_RS00455 (R721_90) | 74109..74285 | - | 177 | WP_000753050 | hypothetical protein | - |
| HXE98_RS00460 (R721_91) | 74278..74493 | - | 216 | WP_001127357 | DUF1187 family protein | - |
| HXE98_RS00465 (R721_92) | 74486..74992 | - | 507 | WP_001326595 | CaiF/GrlA family transcriptional regulator | - |
Host bacterium
| ID | 170 | GenBank | NC_002525 |
| Plasmid name | R721 | Incompatibility group | IncI2 |
| Plasmid size | 75582 bp | Coordinate of oriT [Strand] | 29007..29059 [-] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | dfrA1, ant(3'')-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |