Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100150 |
| Name | oriT_NAH7 |
| Organism | Pseudomonas putida |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_007926 (19758..20187 [-], 430 nt) |
| oriT length | 430 nt |
| IRs (inverted repeats) | IR1: 60..65, 76..81 (GTGTAT..ATACAC) IR2: 121..126, 127..132 (ATACAT..ATGTAT) IR3: 140..144, 151..155 (AAAAA..TTTTT) IR4: 171..175, 179..183 (ACAGC..GCTGT) |
| Location of nic site | 213..214 |
| Conserved sequence flanking the nic site |
_ |
| Note |
oriT sequence
Download Length: 430 nt
GGTGTTCCTCTCTGAGCCCAAAATGGGACATATTCAGGCTAACGATAACTGCCGAAACAGTGTATAGCAAATGTAATACACAAAAAGCTTGATTTTGCTGATCTAGACTGTACTTTTGTAATACATATGTATAGCGCATAAAAACACCACTTTTTCTCTGCAGGCCGCGCACAGCGCGGCTGTCACCTCCCGCTAGGGGGGTGCGTATTGTGTATTGAGACTTTGAATTGAGACTTTGACCCTGTTAAGGGGAAAAGTTGAAGCTCTTGTTAAGGGCGTATCCACTAGCCAGGCCGAAGGCCGTAGGGGGGTAGCACAGATGGTATGCAGAAAGGGTGCTAGAAGAGAGCAGGAAGAGTGCAACTTGACTACAGGTAGGGGGCGAGTAAGATGCACATAGAGTGCAATAGGGGTGCAGATAGGGTGCAAA
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file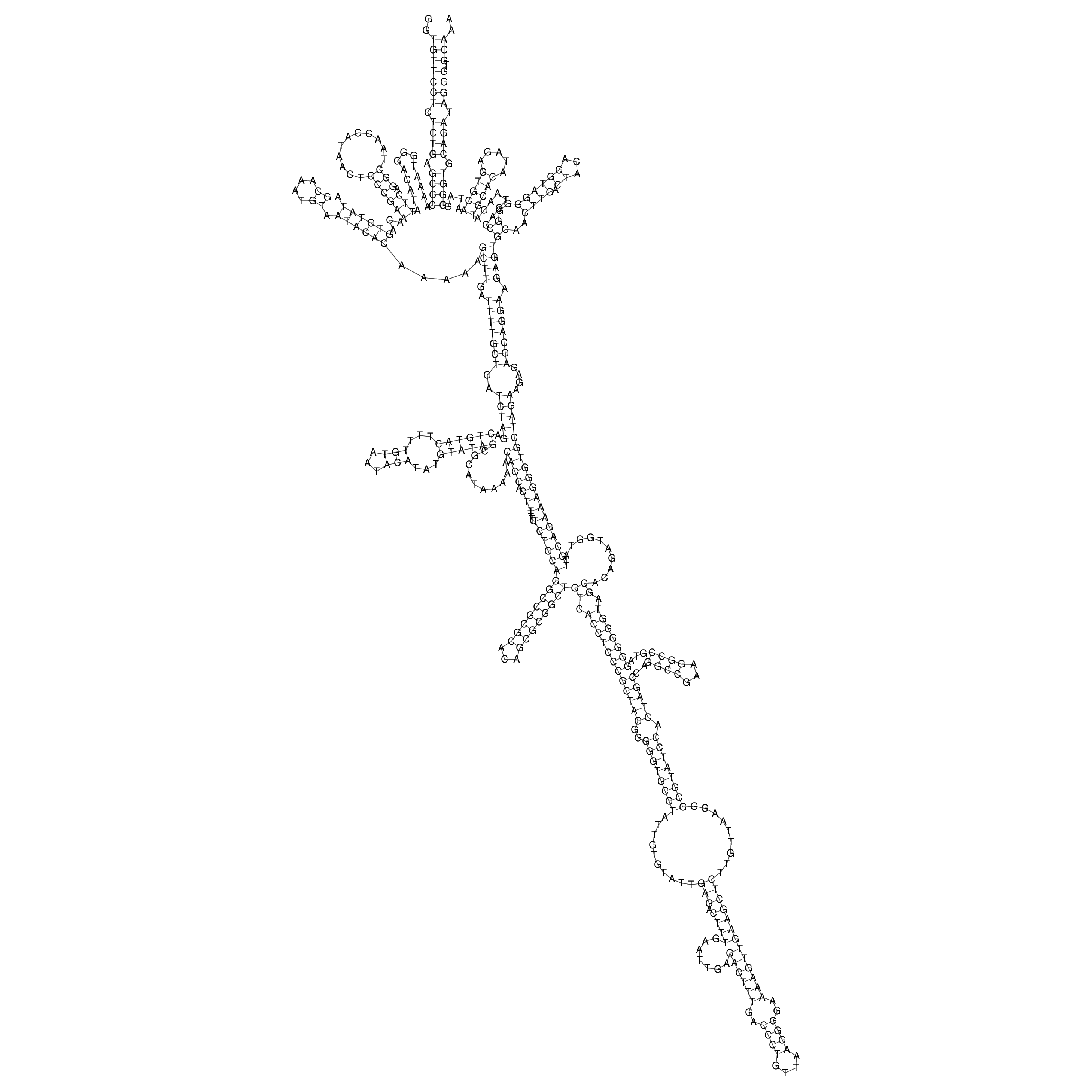
Reference
[1] Kouhei Kishida et al. (2017) Host Range of the Conjugative Transfer System of IncP-9 Naphthalene-Catabolic Plasmid NAH7 and Characterization of Its oriT Region and Relaxase. Applied and environmental microbiology. 83(1):e02359-16. [PMID:27742684]
[2] Sota M et al. (2006) Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol. 188(11):4057-67. [PMID:16707697]
Relaxase
| ID | 144 | GenBank | YP_534815 |
| Name | TraC_NAH7 |
UniProt ID | Q1XGM7 |
| Length | 978 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 978 a.a. Molecular weight: 108592.12 Da Isoelectric Point: 9.9224
MFNVTSIKGSNQYAAAGYYTAADDYYAKESPGEWQGIGAELLGLAGPIDQKELAKLFDGKLPNGEVMPKT
FDKETGNRRMGLDLTFSAPKSVSMQALIAGDKDVVAAHDRAVTKAMEHVEKLAQARRKEHGKSMLERTGN
LIIGKFRHELSRAKDPQLHTHAVVMNATRREDGKWRAIHNDDIFKIQPQIDAMYKGELAKELRELGYEIR
VLDKDGNFELAHISRDQIEAFSSRAKVIEEALAKDGKTRSNATALEKQIIAMATRPRKDERDRHLVKEYW
VTKARDLGIEFGGRSQLDNQEYGRRSESIHAEHNLPEGITAGQAVVQYAINHLTEREQVVGESDLRTAAL
RRAVGLASPSEVDDEIKRLVKQGTLIESPPTYRMATGKDGAALSPAGWRALLKEQKGWSEKEAQQYVKMA
INRGSLVEAEKRYTTQRALKREKAILAIERSGRGQVAPLLSKEQVAKALESSTLSAGQYQAVEVIVSTSN
RFVGIQGDAGTGKTYSVDRAVKLIESVNNAMTERSTTTDAVFRVVALAPYGNQVTALKNEGLDAHTLASF
FHTKDKKLDERTIVVLDEAGVVGARQMEQLMRIIEQSGARLVQLGDTKQTEAIEAGKPFAQLQQNGMQTA
RIKEIQRQKNQELKIAVQHAADGNPGKSLEHVNHVEELREPGQRHQAIVRDYMSLTPQERKEVLIVAGTN
KDRKQINAMTRESLGLVGNGKELPTLNRVDTTQAERRYAPSYKKGMIVQPEKDYIRAGLSRGELYTVDQA
LPGNVLVVKDKNGNRVEFNPRKLTKLSVYNLEKPEFSVGDLVRITRNDQKLDLTNGDRMRVVGNANGVIE
LASLKEKDGTPERVVALPTNRPLHLEHAYSATVHSAQGLTNDRVMISINTKSLTTSQNLWYVAISRARHE
ARIYTDSIAGLPAAIANRYDKTTALSLQQARERQRNESIKPRTVLDGKELERKQRSALDGAGLGKSGV
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q1XGM7 |
Reference
[1] Sota M et al. (2006) Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol. 188(11):4057-67. [PMID:16707697]
Auxiliary protein
| ID | 209 | GenBank | YP_534813 |
| Name | TraA_NAH7 |
UniProt ID | Q1XGM9 |
| Length | 130 a.a. | PDB ID | _ |
| Note | putative plasmid transfer protein TraA | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14469.20 Da Isoelectric Point: 4.6173
MAKTLPIRLNDDQEARLEQAAALAGYKHLSTYVRDRALSNEKSGGRQTDLDSWADQQRMLGLLENVSQAQ
AENRTLLTIAVYLLRQGLQQGKVNELRAELSHLADADQLINTLLPELVSDIERLSGEDNA
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q1XGM9 |
Reference
[1] Sota M et al. (2006) Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol. 188(11):4057-67. [PMID:16707697]
T4CP
| ID | 144 | GenBank | YP_534814 |
| Name | TraB_NAH7 |
UniProt ID | Q1XGM8 |
| Length | 519 a.a. | PDB ID | _ |
| Note | Type IV secretion-system coupling protein DNA-binding domain; pfam10412 | ||
T4CP protein sequence
Download Length: 519 a.a. Molecular weight: 58521.92 Da Isoelectric Point: 8.5125
MHKREVDSAVMTAGFGLPIAGWAAGVYFAQMPLTPFPAAFKETLHNGWHNPIVLGATIATSAVAASLVYY
LYEYCDDGFRGEQYQEFLRGSEIKNWHTVKAKVRRRNEKTNRERKKWGLAKSEPIMIGKLPMPLHLEDRN
TMICASIGAGKSVTMEGMIASALKRGDRLAVVDPNGTFYSKFSFKGDSILNPFDERSVGWTIFNEIRGIH
DFNRMAKSIIPPQVDPSDEQWCAYARDVLSDTMRKLKETNNPDQDTLVNLLVREDGDTIRAFLANTDSEG
YFRDNAEKAIASIQFMMNKYIRPLRYMSKGEFSIYKWVNDPNAGNLFITWREDMRDTMKPLVATWIDTIC
ATILSSEPMTGKRLWLFLDELQSLGKLESFVPAATKGRKHGLRMVGSLQDWSQLNASYGRDDAETLLSCF
RNYVILAAANAKNAGMASEILGSHDVKRWRKSYTAGKLTRTEEVTRDEPVVQASEISNLPDLVAYVKFGE
DFPITKVKTPYIDYKERAQAIMLTGQAAA
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q1XGM8 |
Reference
[1] Sota M et al. (2006) Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol. 188(11):4057-67. [PMID:16707697]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 70997..81295
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HXC10_RS00410 (NAH7_66) | 66544..68031 | - | 1488 | WP_011475407 | SulP family inorganic anion transporter | - |
| HXC10_RS00415 (NAH7_67) | 68550..68846 | - | 297 | WP_011475408 | hypothetical protein | - |
| HXC10_RS00420 (NAH7_68) | 68860..69498 | - | 639 | WP_011475409 | hypothetical protein | - |
| HXC10_RS00425 (NAH7_69) | 69546..70028 | - | 483 | WP_011475410 | hypothetical protein | - |
| HXC10_RS00430 (NAH7_70) | 70043..70648 | - | 606 | WP_011475411 | phospholipase D family protein | - |
| HXC10_RS00435 (NAH7_71) | 70638..70988 | - | 351 | WP_238841818 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HXC10_RS00440 (NAH7_72) | 70997..71902 | - | 906 | WP_011475413 | lytic transglycosylase domain-containing protein | virB1 |
| HXC10_RS00445 (NAH7_73) | 71886..72914 | - | 1029 | WP_011475414 | P-type DNA transfer ATPase VirB11 | virB11 |
| HXC10_RS00450 (NAH7_74) | 72924..74207 | - | 1284 | WP_011475415 | TrbI/VirB10 family protein | virB10 |
| HXC10_RS00455 (NAH7_75) | 74210..75001 | - | 792 | WP_011475416 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| HXC10_RS00460 (NAH7_76) | 75026..75691 | - | 666 | WP_011475417 | type IV secretion system protein | virB8 |
| HXC10_RS00465 (NAH7_77) | 75695..76558 | - | 864 | WP_011475418 | type IV secretion system protein | virB6 |
| HXC10_RS00470 (NAH7_78) | 76546..76950 | - | 405 | WP_011475419 | hypothetical protein | - |
| HXC10_RS00475 (NAH7_79) | 76943..77155 | - | 213 | WP_011475420 | hypothetical protein | - |
| HXC10_RS00480 (NAH7_80) | 77183..77866 | - | 684 | WP_011475421 | type IV secretion system protein | virB5 |
| HXC10_RS00485 (NAH7_81) | 77866..80508 | - | 2643 | WP_151140442 | type VI secretion protein | virb4 |
| HXC10_RS00490 (NAH7_82) | 80537..80830 | - | 294 | WP_011475423 | VirB3 family type IV secretion system protein | virB3 |
| HXC10_RS00495 (NAH7_83) | 80834..81295 | - | 462 | WP_011475424 | TrbC/VirB2 family protein | virB2 |
| HXC10_RS00500 (NAH7_84) | 81312..81587 | - | 276 | WP_238841819 | hypothetical protein | - |
Host bacterium
| ID | 144 | GenBank | NC_007926 |
| Plasmid name | NAH7 | Incompatibility group | IncP9 |
| Plasmid size | 82232 bp | Coordinate of oriT [Strand] | 19758..20187 [-] |
| Host baterium | Pseudomonas putida |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | nahAa, nahAb, nahAc, nahAd, nahB, nahF, nahC, nahE, nahD, nahG, nahH, nahI, nahN, nahL, nahO, nahM, nahK, nahJ |
| Symbiosis gene | - |
| Anti-CRISPR | - |