Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100144 |
| Name | oriT_pTiBo542 |
| Organism | Agrobacterium tumefaciens Ti |
| Sequence Completeness | incomplete |
| NCBI accession of oriT (coordinates [strand]) | DQ058764 (213650..213663 [+], 14 nt) |
| oriT length | 14 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 6..7 |
| Conserved sequence flanking the nic site |
_ |
| Note | Ti plasmid |
oriT sequence
Download Length: 14 nt
CAAGGGCGCAATTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file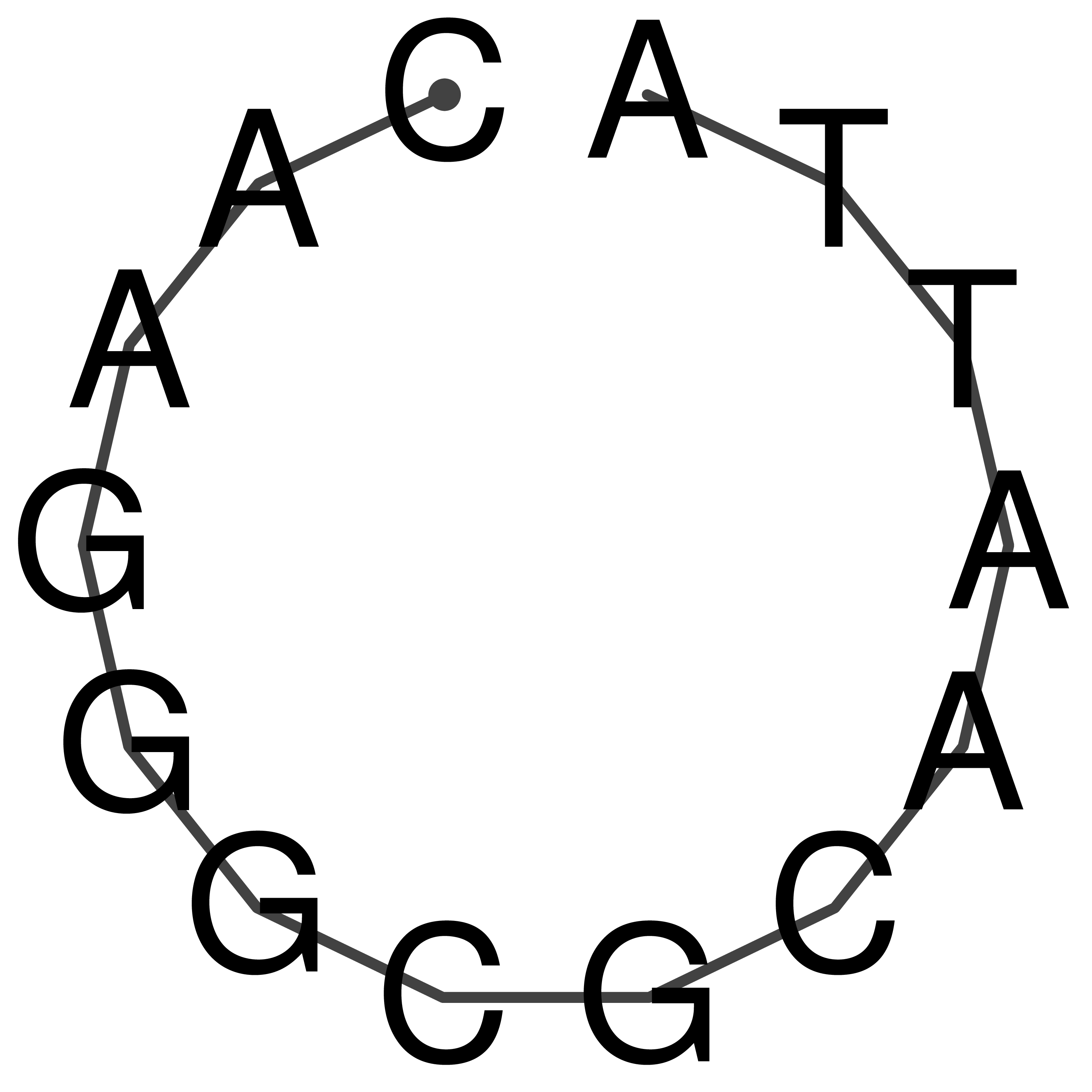
Reference
[1] Wetzel ME et al. (2015) The repABC Plasmids with Quorum-Regulated Transfer Systems in Members of the Rhizobiales Divide into Two Structurally and Separately Evolving Groups. Genome Biol Evol. 7(12):3337-57. [PMID:26590210]
Relaxase
| ID | 138 | GenBank | AAZ50586 |
| Name | TraA_pTiBo542 |
UniProt ID | A5WYB8 |
| Length | 1100 a.a. | PDB ID | |
| Note | Dtr system oriT relaxase; Provisional | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123639.35 Da Isoelectric Point: 10.0980
MAIAHFSASIISRGDGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFTLPADAPKWAKALIADR
AVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIRAKGMVADWVYHDNPGN
PHVHLMSTLRPLSEDGFGSKKVAVIGEDGQPVRTKSGKILYELWGGSTDAFNVLRDGWFERQNHHLALAE
IDLRIDGRSYEKQGIELEPTIHLGVGAKAIERKAASRGVRPELERIELNEERRSENARRILRNPAIVLDL
ITREKSVFDERDIAKVLHRYIDDPAVFQQLMVRIILNPEVLRLQRETIDFATAKKVPARYSTRAMIRLEA
TMARQAMWLSNRELRGVSPSALNATFGRHARLSDEQKAAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLSSEAGIESRTLSSWELRWNRGRDVLDNKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALALYN
ANAGIVGERLKAEAVERLIAHWNHDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFRTADG
ERRFDAGDQIVFLKNETSLGVKNGMIGHVVEAAPNRIVAVVGDRDHRRHVVVEQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAENRGLHIMQVARTMLRDRLDWTLRQKTKVSDLVHRLRALGERLGPDQSPKTQTMKEAGPMVAG
IKTFSGSVADTVGDKLGADPTLKQQWEEVSARFRYVFADPVTAFRAINFDTVLADKEMAKQVLQKLEAEP
ETIGPLKGKTGILAGKAEREARRIAEVNVPALKRDLEQYLQMRETATMRHQADEQTLRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFSQSVTERFGERTLLNNAAREPSGTLYEKLSEG
MTPEQKQQLKQAWPVMRTAQQLAAHERTAQSLRQAEEQRLTQRQVPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A5WYB8 |
Auxiliary protein
| ID | 202 | GenBank | AAZ50585 |
| Name | TraC_pTiBo542 |
UniProt ID | A5WYB7 |
| Length | 98 a.a. | PDB ID | _ |
| Note | conjugal transfer protein TraC | ||
Auxiliary protein sequence
Download Length: 98 a.a. Molecular weight: 10208.37 Da Isoelectric Point: 6.4042
MKKPSSKIREEIARLQDQLKQAETREAERIGRIALKAGLGEIEIDEADLQAALEEIAKRFRGGKGIATGK
KDSGDGRNASGASTAQSSGADAGGIGEA
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A5WYB7 |
| ID | 203 | GenBank | AAZ50584 |
| Name | TraD_pTiBo542 |
UniProt ID | A5WYB6 |
| Length | 71 a.a. | PDB ID | _ |
| Note | conjugal transfer protein TraD | ||
Auxiliary protein sequence
Download Length: 71 a.a. Molecular weight: 7798.99 Da Isoelectric Point: 10.1998
MAKTITSDARKKDTREKIELGGLIVKAGLRYEKRSLLLGLLIDANRRIRGDETEKARLAAIGAEAFGNEG
E
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A5WYB6 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 90008..99663
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| pTiBo070 | 85029..86795 | + | 1767 | AAZ50458 | MoaD | - |
| pTiBo071 | 86914..88482 | + | 1569 | AAZ50459 | MoaA | - |
| pTiBo072 | 88652..89047 | - | 396 | AAZ50460 | orf_Bo072 | - |
| pTiBo073 | 89084..89737 | + | 654 | AAZ50461 | orf_Bo073 | - |
| pTiBo074 | 90008..91312 | - | 1305 | AAZ50462 | TrbI | virB10 |
| pTiBo075 | 91325..91801 | - | 477 | AAZ50463 | TrbH | - |
| pTiBo076 | 91801..92667 | - | 867 | AAZ50464 | TrbG | virB9 |
| pTiBo077 | 92673..93335 | - | 663 | AAZ50465 | TrbF | virB8 |
| pTiBo078 | 93350..94537 | - | 1188 | AAZ50466 | TrbL | virB6 |
| pTiBo079 | 94531..94749 | - | 219 | AAZ50467 | TrbK | - |
| pTiBo080 | 94746..95555 | - | 810 | AAZ50468 | TrbJ | virB5 |
| pTiBo081 | 95527..97995 | - | 2469 | AAZ50469 | TrbE | virb4 |
| pTiBo082 | 98006..98305 | - | 300 | AAZ50470 | TrbD | virB3 |
| pTiBo083 | 98298..98702 | - | 405 | AAZ50471 | TrbC | virB2 |
| pTiBo084 | 98692..99663 | - | 972 | AAZ50472 | TrbB | virB11 |
| pTiBo085 | 99660..100298 | - | 639 | AAZ50473 | TraI | - |
| pTiBo086 | 100664..101860 | + | 1197 | AAZ50474 | RepA | - |
| pTiBo087 | 101963..102922 | + | 960 | AAZ50475 | RepB | - |
| pTiBo088 | 103097..104311 | + | 1215 | AAZ50476 | RepC | - |
Region 2: 153093..162528
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| pTiBo128 | 148219..148605 | + | 387 | AAZ50513 | orf_Bo128 | - |
| pTiBo129 | 148602..148946 | + | 345 | AAZ50514 | orf_Bo129 | - |
| pTiBo130 | 149021..150610 | + | 1590 | AAZ50515 | orf_Bo130 | - |
| pTiBo131 | 150873..151616 | + | 744 | AAZ50516 | VirJ | - |
| pTiBo132 | 151620..151997 | - | 378 | AAZ50517 | orf_Bo132 | - |
| pTiBo133 | 153093..153812 | + | 720 | AAZ50518 | VirB1 | virB1 |
| pTiBo134 | 153827..154192 | + | 366 | AAZ50519 | VirB2 | virB2 |
| pTiBo135 | 154192..154518 | + | 327 | AAZ50520 | VirB3 | virB3 |
| pTiBo136 | 154518..156887 | + | 2370 | AAZ50521 | VirB4 | virb4 |
| pTiBo137 | 156902..157564 | + | 663 | AAZ50522 | VirB5 | - |
| pTiBo138 | 157661..158548 | + | 888 | AAZ50523 | VirB6 | virB6 |
| pTiBo139 | 158582..158749 | + | 168 | AAZ50524 | VirB7 | - |
| pTiBo140 | 158736..159449 | + | 714 | AAZ50525 | VirB8 | virB8 |
| pTiBo141 | 159446..160327 | + | 882 | AAZ50526 | VirB9 | virB9 |
| pTiBo142 | 160324..161457 | + | 1134 | AAZ50527 | VirB10 | virB10 |
| pTiBo143 | 161497..162528 | + | 1032 | AAZ50528 | VirB11 | virB11 |
| pTiBo144 | 162660..163463 | + | 804 | AAZ50529 | VirG | - |
| pTiBo145 | 163574..164182 | - | 609 | AAZ50530 | VirC2 | - |
| pTiBo146 | 164185..164880 | - | 696 | AAZ50531 | VirC1 | - |
| pTiBo147 | 165149..165592 | + | 444 | AAZ50532 | VirD1 | - |
| pTiBo148 | 165626..166900 | + | 1275 | AAZ50533 | VirD2 | - |
Host bacterium
| ID | 138 | GenBank | DQ058764 |
| Plasmid name | pTiBo542 | Incompatibility group | _ |
| Plasmid size | 244978 bp | Coordinate of oriT [Strand] | 213650..213663 [+] |
| Host baterium | Agrobacterium tumefaciens Ti |
Cargo genes
| Drug resistance gene | _ |
| Virulence gene | virulence-tumorigenic |
| Metal resistance gene | _ |
| Degradation gene | _ |
| Symbiosis gene | - |
| Anti-CRISPR | - |