Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100140 |
| Name | oriT2_pMAS2027 |
| Organism | Escherichia coli |
| Sequence Completeness | incomplete |
| NCBI accession of oriT (coordinates [strand]) | NC_013503 ( 38362..38380 [+], 19 nt) |
| oriT length | 19 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 13..14 |
| Conserved sequence flanking the nic site |
TATCCTG|C |
| Note | _ |
oriT sequence
Download Length: 19 nt
TTGCCCTATCCTGCATCGC
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file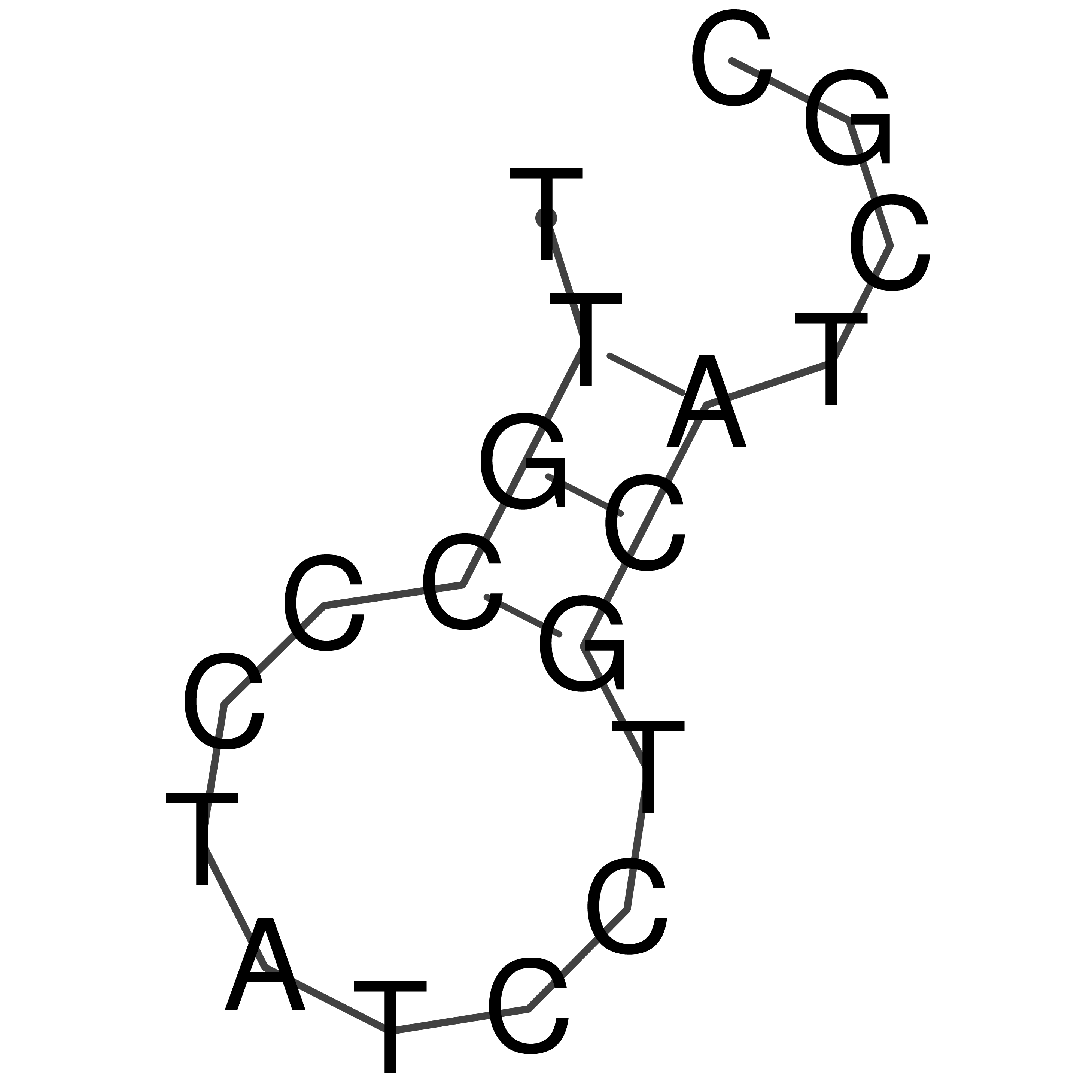
Reference
[1] Ong CL et al. (2009) Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 75(21):6783-91. [PMID:19717626]
Relaxase
| ID | 134 | GenBank | YP_003292208 |
| Name | TaxC_pMAS2027 |
UniProt ID | D0QMQ2 |
| Length | 390 a.a. | PDB ID | |
| Note | Relaxase | ||
Relaxase protein sequence
Download Length: 390 a.a. Molecular weight: 44752.18 Da Isoelectric Point: 10.5067
MIMGVYVDKEYRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAVTRQGIRN
SIDYMSRESELPVMSESGRVWTGDEILEAKDHMIDRANDPQHVMNDKGKENKKITQNIVFSPPVSAKVKP
EDLLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLKG
YDVKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDVGYDHYQNDKTKSKQHFIKLKTLNKGVEKTYWGA
DFGDLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGVDRTPSASKELV
LNSPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFWSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D0QMQ2 |
Reference
[1] Ong CL et al. (2009) Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 75(21):6783-91. [PMID:19717626]
Auxiliary protein
| ID | 197 | GenBank | YP_003292209 |
| Name | Ddp1_pMAS2027 |
UniProt ID | D0QMQ3 |
| Length | 204 a.a. | PDB ID | _ |
| Note | similar to TaxA from R6K | ||
Auxiliary protein sequence
Download Length: 204 a.a. Molecular weight: 24091.59 Da Isoelectric Point: 8.0185
MTSMTPFRNVCPVTYSRFFLENFMKKVQFRIDENQHDDLLDCLKTLYPDEPALTVAKGMKLLANALLKSK
AGSKDINTFFDNNDFIKTTMYLTGKQRADIERAANRHGWTLSRECRYRIQTTLENELDFFDQELLMMNRC
RNSIDKIGRNFHYIIVNDQTRVLDKDGFYQDAERLTTEIFNLKNQFENYIMLCKGRTVSNKVEM
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D0QMQ3 |
Reference
[1] Ong CL et al. (2009) Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 75(21):6783-91. [PMID:19717626]
T4CP
| ID | 134 | GenBank | YP_003292195 |
| Name | TaxB_pMAS2027 |
UniProt ID | D0QMN9 |
| Length | 613 a.a. | PDB ID | _ |
| Note | bacterial conjugative protein involved in type IV sectretion | ||
T4CP protein sequence
Download Length: 613 a.a. Molecular weight: 69645.64 Da Isoelectric Point: 7.1491
MIMSLKLPDKGQWAFIGLVMCLVTYYTGSVAVYFLNGKTPLYIWKNFDSMLLWRIITESNIRIDIRLTAI
PSLLSGMVSSLIVPVFIIWQLNKTDVALYGDAKFASDNDLRKSKLLKWEKENDTDILVGAYKGKYLWYTA
PDFVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNP
LFYIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVF
SIGSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLF
YLPTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLM
FLDEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAK
ISEKLGYITTTSKSTSKSRGRSTSQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLD
PYFMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D0QMN9 |
Reference
[1] Ong CL et al. (2009) Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 75(21):6783-91. [PMID:19717626]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 25562..35522
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS641_RS00130 (pMAS2027_32) | 21935..22900 | + | 966 | WP_001328098 | SMEK domain-containing protein | - |
| HS641_RS00135 (pMAS2027_33) | 23015..23317 | - | 303 | WP_000717624 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HS641_RS00140 (pMAS2027_34) | 23314..23727 | - | 414 | WP_000722603 | cag pathogenicity island Cag12 family protein | - |
| HS641_RS00145 (pMAS2027_35) | 23724..25559 | - | 1836 | WP_014106755 | type IV secretory system conjugative DNA transfer family protein | - |
| HS641_RS00150 (pMAS2027_36) | 25562..26593 | - | 1032 | WP_001058465 | P-type DNA transfer ATPase VirB11 | virB11 |
| HS641_RS00155 (pMAS2027_37) | 26595..27800 | - | 1206 | WP_000139139 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| HS641_RS00160 (pMAS2027_38) | 27797..28726 | - | 930 | WP_000783379 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| HS641_RS00165 (pMAS2027_39) | 28731..29444 | - | 714 | WP_000394615 | type IV secretion system protein | virB8 |
| HS641_RS00270 (pMAS2027_40) | 29434..29562 | - | 129 | WP_001328099 | hypothetical protein | - |
| HS641_RS00170 (pMAS2027_41) | 29689..30822 | - | 1134 | WP_000513457 | type IV secretion system protein | virB6 |
| HS641_RS00175 (pMAS2027_42) | 30832..31059 | - | 228 | WP_000835349 | EexN family lipoprotein | - |
| HS641_RS00180 (pMAS2027_43) | 31060..31818 | - | 759 | WP_000744200 | type IV secretion system protein | - |
| HS641_RS00185 (pMAS2027_44) | 31829..34582 | - | 2754 | WP_001352869 | VirB3 family type IV secretion system protein | virb4 |
| HS641_RS00190 (pMAS2027_45) | 34601..34897 | - | 297 | WP_000916182 | TrbC/VirB2 family protein | virB2 |
| HS641_RS00195 (pMAS2027_46) | 34875..35522 | - | 648 | WP_000953526 | lytic transglycosylase domain-containing protein | virB1 |
| HS641_RS00200 | 35555..35767 | - | 213 | WP_000125172 | hypothetical protein | - |
| HS641_RS00205 (pMAS2027_47) | 35697..36215 | - | 519 | WP_001446885 | transcription termination/antitermination NusG family protein | - |
| HS641_RS00210 (pMAS2027_48) | 36568..37734 | - | 1167 | WP_000539538 | MobP1 family relaxase | - |
| HS641_RS00215 (pMAS2027_49) | 37737..38282 | - | 546 | WP_001426313 | DNA distortion polypeptide 1 | - |
| HS641_RS00225 (pMAS2027_51) | 38608..38880 | - | 273 | WP_000167280 | hypothetical protein | - |
| HS641_RS00230 | 38897..39091 | - | 195 | WP_001714231 | hypothetical protein | - |
| HS641_RS00275 | 39113..39292 | - | 180 | WP_024133577 | hypothetical protein | - |
| HS641_RS00235 (pMAS2027_52) | 39335..39664 | - | 330 | WP_000866654 | hypothetical protein | - |
| HS641_RS00240 (pMAS2027_53) | 39758..40000 | - | 243 | WP_000008708 | hypothetical protein | - |
| HS641_RS00245 (pMAS2027_54) | 39990..40271 | - | 282 | WP_012845398 | hypothetical protein | - |
Host bacterium
| ID | 134 | GenBank | NC_013503 |
| Plasmid name | pMAS2027 | Incompatibility group | IncX1 |
| Plasmid size | 42644 bp | Coordinate of oriT [Strand] | 2060..2078 [-]; 38362..38380 [+] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | mrkF, mrkD, mrkC, mrkB, mrkA |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |