Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100116 |
| Name | oriT_pSC101 |
| Organism | Salmonella enterica subsp. enterica serovar Typhimurium |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_002056 (4071..4115 [+], 45 nt) |
| oriT length | 45 nt |
| IRs (inverted repeats) | 10..18, 22..30 (TTTCTGAAC.. GTGAAGAAA) |
| Location of nic site | 41..42 |
| Conserved sequence flanking the nic site |
TAAGTGCG|CCCT |
| Note | IncQ type mobilizable plasmid |
oriT sequence
Download Length: 45 nt
GGGCGCACGTTTCTGAACGAAGTGAAGAAAGTCTAAGTGCGCCCT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file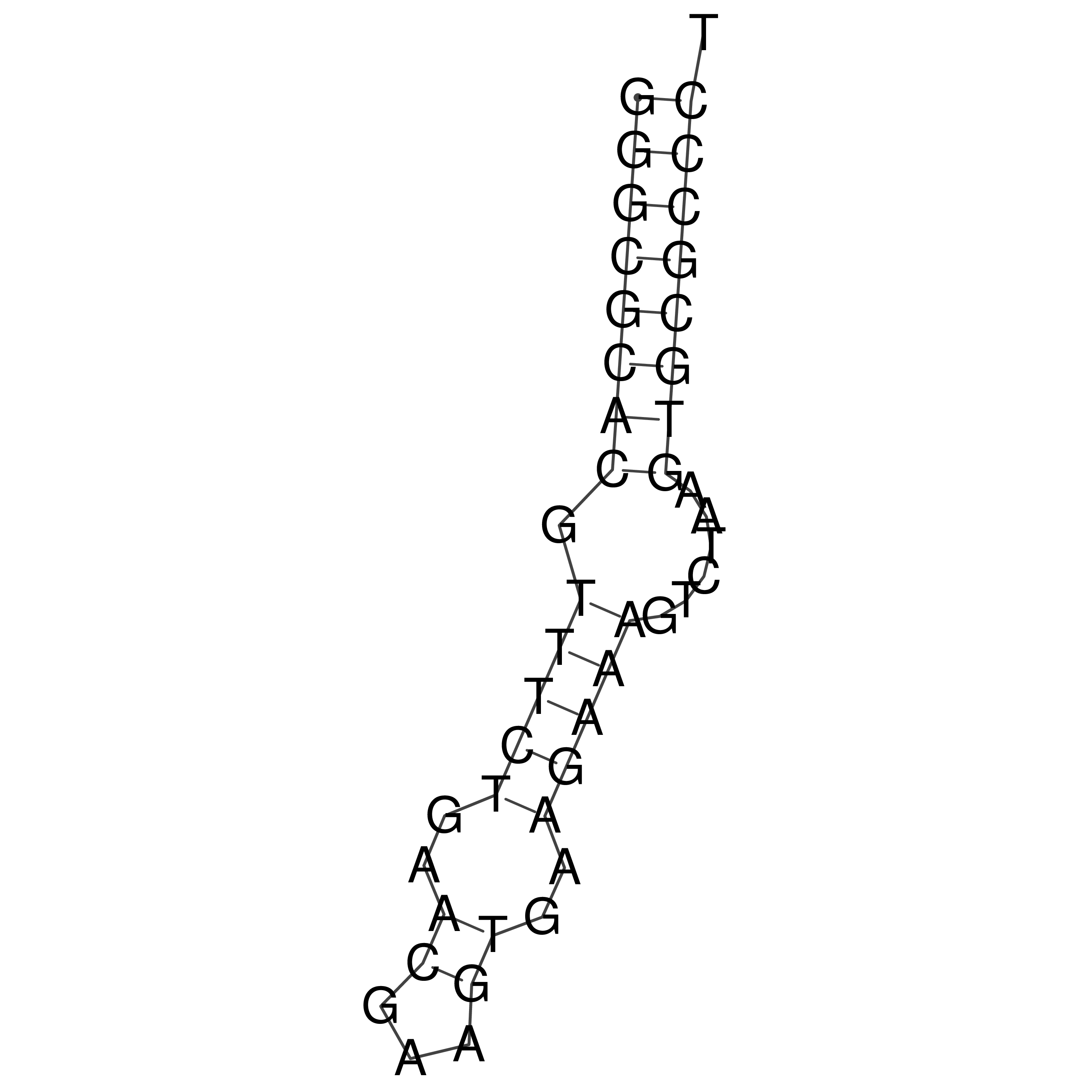
Reference
[1] Jandle S et al. (2006) Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J Bacteriol. 188(2):499-506. [PMID:16385040]
[2] Parker C et al. (2005) Elements in the co-evolution of relaxases and their origins of transfer. Plasmid . 53(2):113-8. [PMID:15737398]
[3] Meyer R (2000) Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J Bacteriol. 182(17):4875-81. [PMID:10940031]
Relaxase
| ID | 111 | GenBank | NP_044284 |
| Name | MobA_pSC101 |
UniProt ID | P14492 |
| Length | 371 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 371 a.a. Molecular weight: 43086.75 Da Isoelectric Point: 6.9522
MASYHLSVKTGGKGSASPHADYIAREGKYAREKDSDLEHKESGNMPAWAAHKPSEFWKAADTSERANGCT
YREIEIALPRELKPEQRLELVRDFVQQEIGDRHAYQFAIHNPKAAIAGGEQPHAHIMFSERINDGIHRDP
EQYFKRANTKEPDAVAQKRHVSGKHRPNAKNTLLPRGRRWADLQNKHLERYQHADRVDSRSLKAQGIDRE
PERHLGAGQVQRFDTEQLQAILERREAERQVQQSRDERDSVIDVTTSLREALSERDTLTLKQELKSEPEQ
ESHSGRTFDFEKEPDKLNALVSDAMKDIQEEIDLQSLVNDAMAEFQGIHQEMERQRERERLAEKQRQQEK
ERQRLAEQIRQKPDKGWSFSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P14492 |
Reference
[1] Becker EC et al. (2012) Origin and fate of the 3' ends of single-stranded DNA generated by conjugal transfer of plasmid R1162. J Bacteriol. 194(19):5368-76. [PMID:22865840]
[2] Jandle S et al. (2006) Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J Bacteriol. 188(2):499-506. [PMID:16385040]
[3] Meyer R (2000) Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J Bacteriol. 182(17):4875-81. [PMID:10940031]
Auxiliary protein
| ID | 167 | GenBank | NP_044283 |
| Name | MobX_pSC101 |
UniProt ID | P14493 |
| Length | 194 a.a. | PDB ID | _ |
| Note | |||
Auxiliary protein sequence
Download Length: 194 a.a. Molecular weight: 21886.09 Da Isoelectric Point: 8.6860
MSDEIKQIAALIGHHQALEKRVTSLTEQFQAASSQLQQQSETLSRVIRELDSASGNMTDTVRKSVSSALT
QVEKELKQAGLAQQKPATEALNQAADTAKAMIHEMRREMSRYTWKSAIYLVLTIFFVLASCVTAFTWFMN
DGYSQIAEMQRMEAVWQKKAPLADISRCDGKPCVKVDTRTTYGDKENTWMIIKK
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P14493 |
Reference
[1] Becker EC et al. (2012) Origin and fate of the 3' ends of single-stranded DNA generated by conjugal transfer of plasmid R1162. J Bacteriol. 194(19):5368-76. [PMID:22865840]
[2] Jandle S et al. (2006) Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J Bacteriol. 188(2):499-506. [PMID:16385040]
Host bacterium
| ID | 111 | GenBank | NC_002056 |
| Plasmid name | pSC101 | Incompatibility group | ColE10 |
| Plasmid size | 9263 bp | Coordinate of oriT [Strand] | 4071..4115 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Typhimurium |
Cargo genes
| Drug resistance gene | tet(C) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |