Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100100 |
| Name | oriT_pTF4.1 |
| Organism | Acidithiobacillus ferrooxidans |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_001520 (245..319 [-], 75 nt) |
| oriT length | 75 nt |
| IRs (inverted repeats) | IR1:3..7, 19..23 (CCTTG..CAAGG) IR2: 49..58, 67..75 (CCACGGGGGA..TCCCCCTGG) |
| Location of nic site | 29..30nt |
| Conserved sequence flanking the nic site |
_ |
| Note | _ |
oriT sequence
Download Length: 75 nt
>oriT_pTF4.1
CTCCTTGCTCGGTTGGGTCAAGGGGCACCGCGCCCTTGCCTTGGGGGTCCACGGGGGAGCGGAGCATCCCCCTGG
CTCCTTGCTCGGTTGGGTCAAGGGGCACCGCGCCCTTGCCTTGGGGGTCCACGGGGGAGCGGAGCATCCCCCTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file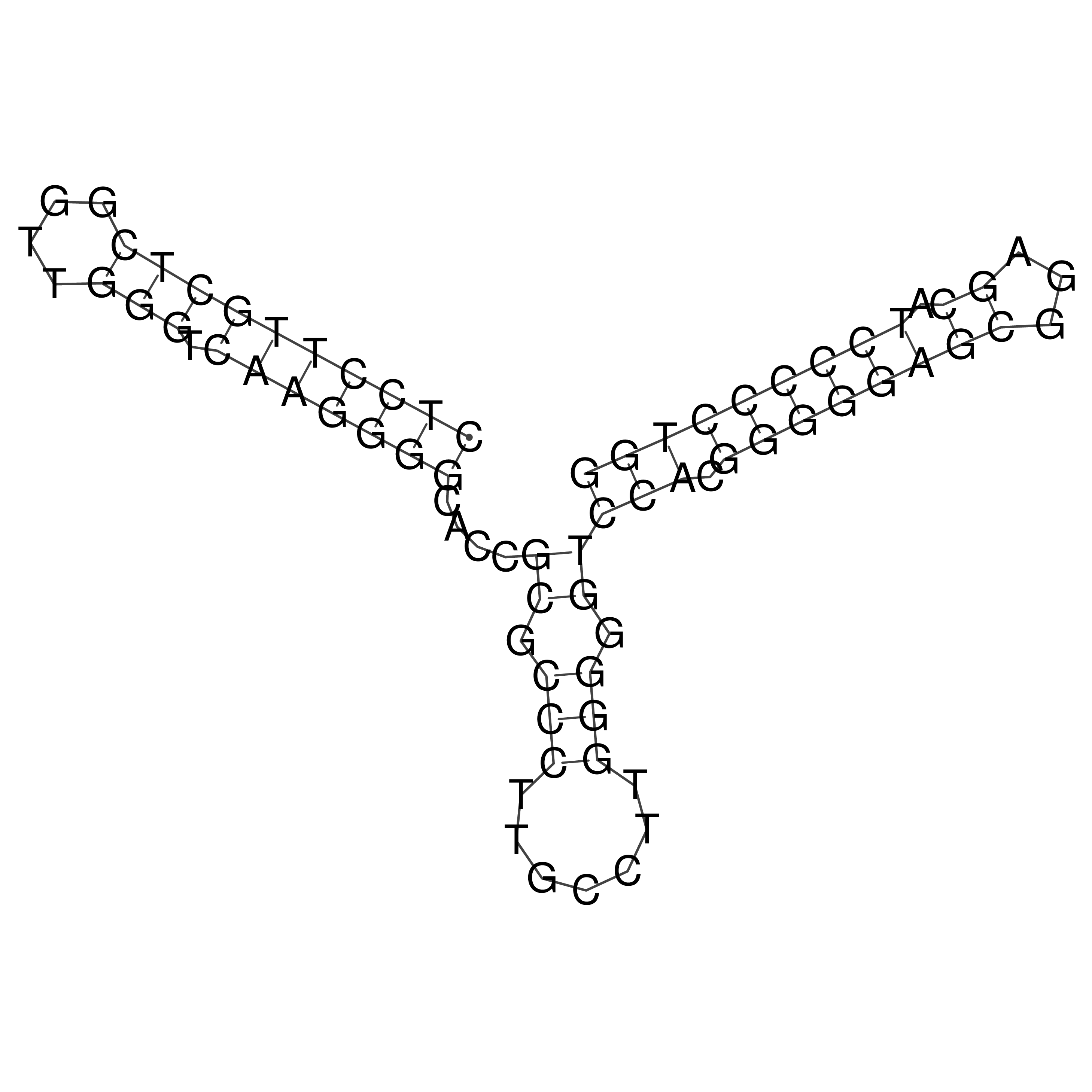
Relaxase
| ID | 95 | GenBank | NP_052137 |
| Name | ORF1_pTF4.1 |
UniProt ID | Q56294 |
| Length | 542 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 542 a.a. Molecular weight: 61320.55 Da Isoelectric Point: 7.1304
>NP_052137.1 unnamed protein product (plasmid) [Acidithiobacillus ferrooxidans]
MEAFGAGTRCKSPLYHANIDPRADERLTPEQWGRAVDALEAKLGFTGQPRALVQHVKEGREHVHVVWSRI
DIERNRAIPDSHNYRKHEEVARALEREFGHERVQGAHVEREGKERPARTPSHAEMQQAERSGLDPKAIKA
EITALWRQTDSGKAFAGALAEAGYILARGDKRDFIVIDQAGESHSLARRIDGVKAADMRARLSDIDQATL
PDARGARAEQAQKALDDAQKAEEARTAEAREAAQKAAEAVERADEADLTRWRSMPLNALQWEINTLRPDR
YAALAAGQAAEPTYRETLEATDAAHAGAVKARGEALEATETVRSLKQKAERYREYQGLLGRVRFWLQDKG
ILEIPEWKRLQTEISKAQKLADKRQNDLATAEALHTEAKSALKTLDSAIAGAVDVAQAPEWRRYEVARSV
LNERIEEDRRRAAEAEAERRAEEIRRRVDAEIEREKREGIEQQRRAEISKIPTIVRGLASITQTEQGTFR
VRYPDLPSEELKRQQDLLRRYPDEAKSAVKAEFSERQVQAKREKDRGFDMMD
MEAFGAGTRCKSPLYHANIDPRADERLTPEQWGRAVDALEAKLGFTGQPRALVQHVKEGREHVHVVWSRI
DIERNRAIPDSHNYRKHEEVARALEREFGHERVQGAHVEREGKERPARTPSHAEMQQAERSGLDPKAIKA
EITALWRQTDSGKAFAGALAEAGYILARGDKRDFIVIDQAGESHSLARRIDGVKAADMRARLSDIDQATL
PDARGARAEQAQKALDDAQKAEEARTAEAREAAQKAAEAVERADEADLTRWRSMPLNALQWEINTLRPDR
YAALAAGQAAEPTYRETLEATDAAHAGAVKARGEALEATETVRSLKQKAERYREYQGLLGRVRFWLQDKG
ILEIPEWKRLQTEISKAQKLADKRQNDLATAEALHTEAKSALKTLDSAIAGAVDVAQAPEWRRYEVARSV
LNERIEEDRRRAAEAEAERRAEEIRRRVDAEIEREKREGIEQQRRAEISKIPTIVRGLASITQTEQGTFR
VRYPDLPSEELKRQQDLLRRYPDEAKSAVKAEFSERQVQAKREKDRGFDMMD
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q56294 |
Auxiliary protein
| ID | 136 | GenBank | NP_052136 |
| Name | ORF4_pTF4.1 |
UniProt ID | Q56293 |
| Length | 121 a.a. | PDB ID | _ |
| Note | Bacterial mobilisation protein (MobC); pfam05713 | ||
Auxiliary protein sequence
Download Length: 121 a.a. Molecular weight: 12524.27 Da Isoelectric Point: 11.0590
>NP_052136.1 unnamed protein product (plasmid) [Acidithiobacillus ferrooxidans]
MASGSETRQRQAALRIRLTAAEHATITAASERAGLSLAGYARSQLLSAPPVRQARRPPVERAELARLLAE
LGKIGSNVNQIAHALNGGGDAVPSDLASVQADIAAIRAAIMLALGREPGEP
MASGSETRQRQAALRIRLTAAEHATITAASERAGLSLAGYARSQLLSAPPVRQARRPPVERAELARLLAE
LGKIGSNVNQIAHALNGGGDAVPSDLASVQADIAAIRAAIMLALGREPGEP
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q56293 |
Host bacterium
| ID | 95 | GenBank | NC_001520 |
| Plasmid name | pTF4.1 | Incompatibility group | - |
| Plasmid size | 4104 bp | Coordinate of oriT [Strand] | 245..319 [-] |
| Host baterium | Acidithiobacillus ferrooxidans |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |