Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100099 |
| Name | oriT_pKPC-LKEc |
| Organism | Escherichia coli strain LK-NARMP |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | KC788405 (30139..30221 [-], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | IR1: 6..13, 17..24 (GTCGGGGC..GCCCTGAC) IR2: 31..47, 54..70 (GTAATTGTAATAGCGTC..GACGGTATTACAATTAC) |
| Location of nic site | 78..79 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | _ |
oriT sequence
Download Length: 83 nt
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file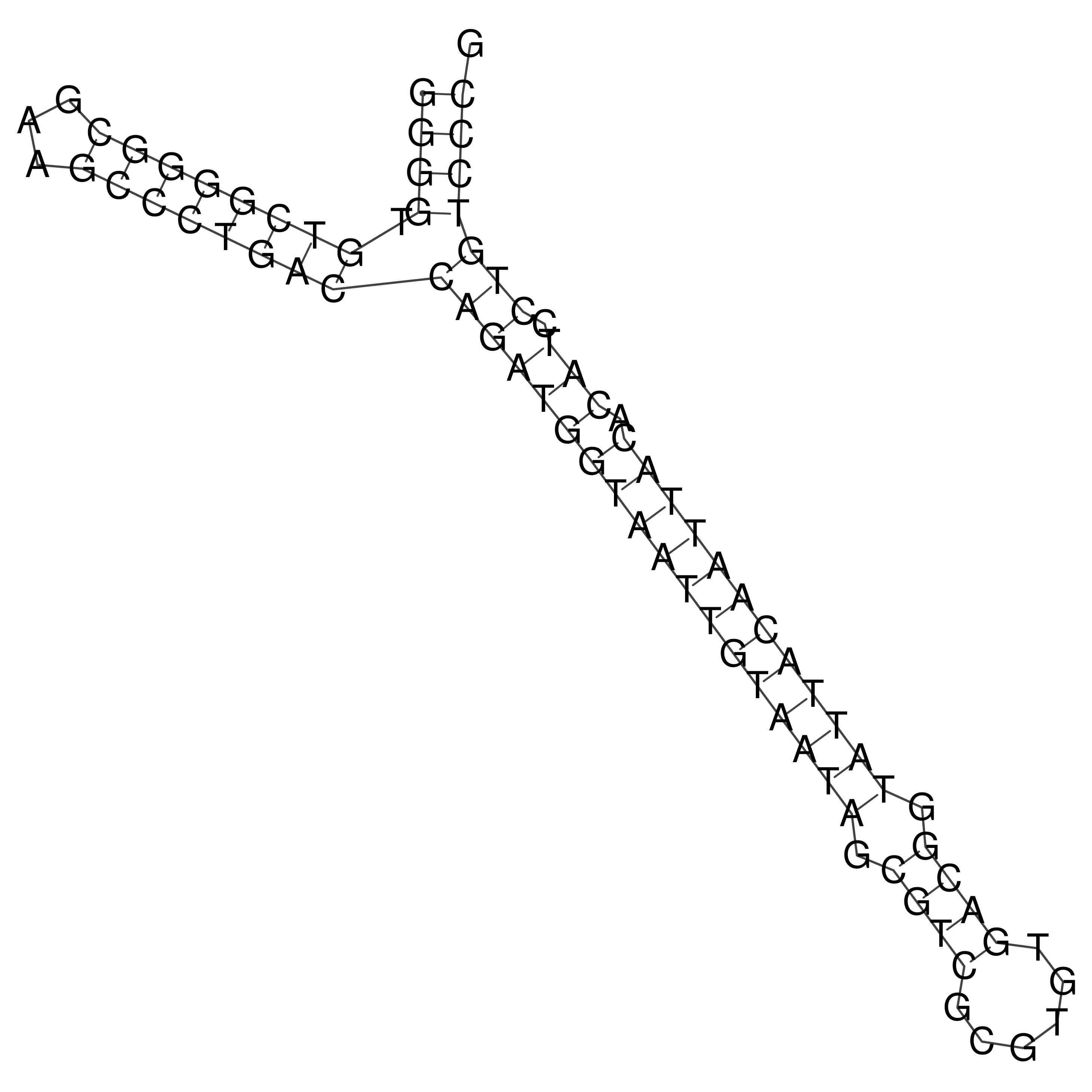
Reference
[1] Chen YT et al. (2014) KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 69(3):628-31. [PMID:24123430]
Relaxase
| ID | 94 | GenBank | AGS77392 |
| Name | NikB_pKPC-LKEc |
UniProt ID | U5KKC0 |
| Length | 899 a.a. | PDB ID | |
| Note | Relaxase/Mobilisation nuclease | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103882.34 Da Isoelectric Point: 7.4198
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | U5KKC0 |
Reference
[1] Chen YT et al. (2014) KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 69(3):628-31. [PMID:24123430]
Auxiliary protein
| ID | 135 | GenBank | AGS77391 |
| Name | NikA_pKPC-LKEc |
UniProt ID | C7SA35 |
| Length | 110 a.a. | PDB ID | _ |
| Note | conjugal transfer relaxosome component | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | C7SA35 |
Reference
[1] Chen YT et al. (2014) KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 69(3):628-31. [PMID:24123430]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 103098..136095
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KPCE_126 | 98250..99512 | - | 1263 | AGS77471 | TnpA | - |
| KPCE_127 | 99665..100045 | - | 381 | AGS77472 | hypothetical protein | - |
| KPCE_128 | 100604..100855 | - | 252 | AGS77473 | hypothetical protein | - |
| KPCE_129 | 101209..101451 | + | 243 | AGS77474 | hypothetical protein | - |
| KPCE_130 | 101516..101692 | - | 177 | AGS77475 | hypothetical protein | - |
| KPCE_131 | 102365..103027 | - | 663 | AGS77476 | ExcB | - |
| KPCE_132 | 102365..102808 | - | 444 | AGS77477 | ExcA | - |
| KPCE_133 | 103098..105266 | - | 2169 | AGS77478 | TraY | traY |
| KPCE_134 | 105363..105947 | - | 585 | AGS77479 | TraX | - |
| KPCE_135 | 105976..107055 | - | 1080 | AGS77480 | TraW | traW |
| KPCE_136 | 107145..107570 | - | 426 | AGS77481 | TraV | traV |
| KPCE_137 | 107759..110803 | - | 3045 | AGS77482 | TraV | traU |
| KPCE_138 | 110893..111693 | - | 801 | AGS77483 | TraU | traT |
| KPCE_139 | 111677..111865 | - | 189 | AGS77484 | TraS | - |
| KPCE_140 | 111929..112333 | - | 405 | AGS77485 | TraR | traR |
| KPCE_141 | 112384..112911 | - | 528 | AGS77486 | TraQ | traQ |
| KPCE_142 | 112911..113615 | - | 705 | AGS77487 | TraP | traP |
| KPCE_143 | 113615..114904 | - | 1290 | AGS77488 | TraO | traO |
| KPCE_144 | 114907..115890 | - | 984 | AGS77489 | TraN | traN |
| KPCE_145 | 115901..116593 | - | 693 | AGS77490 | TraM | traM |
| KPCE_146 | 116590..116937 | - | 348 | AGS77491 | TraL | traL |
| KPCE_147 | 116955..120722 | - | 3768 | AGS77492 | SogL | - |
| KPCE_148 | 116955..119489 | - | 2535 | AGS77493 | SogS | - |
| KPCE_149 | 120812..121363 | - | 552 | AGS77494 | Nuc endonuclease | - |
| KPCE_150 | 121378..121668 | - | 291 | AGS77495 | TraK | traK |
| KPCE_151 | 121665..122813 | - | 1149 | AGS77496 | TraJ | virB11 |
| KPCE_152 | 122810..123628 | - | 819 | AGS77497 | TraI | traI |
| KPCE_153 | 123625..124083 | - | 459 | AGS77498 | TraH | - |
| KPCE_154 | 124478..125062 | - | 585 | AGS77499 | TraG | - |
| KPCE_155 | 125122..126324 | - | 1203 | AGS77500 | TraF | - |
| KPCE_156 | 126410..127234 | - | 825 | AGS77501 | TraE | traE |
| KPCE_157 | 127385..128539 | - | 1155 | AGS77502 | Rci recombinase | - |
| KPCE_158 | 130179..131615 | - | 1437 | AGS77503 | PilV | - |
| KPCE_159 | 131603..132079 | - | 477 | AGS77504 | leader peptidase/N-methyltransferase | - |
| KPCE_160 | 132294..132803 | - | 510 | AGS77505 | PilT | virB1 |
| KPCE_161 | 132813..133427 | - | 615 | AGS77506 | PilS | - |
| KPCE_162 | 133444..134529 | - | 1086 | AGS77507 | PilR | - |
| KPCE_163 | 134542..136095 | - | 1554 | AGS77508 | PilQ ATPase | virB11 |
| KPCE_164 | 136106..136558 | - | 453 | AGS77509 | PilP | - |
| KPCE_165 | 136545..137840 | - | 1296 | AGS77510 | PolO | - |
| KPCE_166 | 137833..139515 | - | 1683 | AGS77511 | PilN lipoprotein | - |
| KPCE_167 | 139529..139966 | - | 438 | AGS77512 | PilM | - |
| KPCE_168 | 139966..141033 | - | 1068 | AGS77513 | PilL | - |
Host bacterium
| ID | 94 | GenBank | KC788405 |
| Plasmid name | pKPC-LKEc | Incompatibility group | IncI |
| Plasmid size | 145401 bp | Coordinate of oriT [Strand] | 30139..30221 [-] |
| Host baterium | Escherichia coli strain LK-NARMP |
Cargo genes
| Drug resistance gene | blaKPC-2 (encoding beta-lactamase KPC-2) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |