Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100097 |
| Name | oriT_pHH2-227 |
| Organism | Uncultured bacterium |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | JN581942 (_) |
| oriT length | 341 nt |
| IRs (inverted repeats) | IR1: 66..78, 85..97 (AACATCTGTGGTC..GACCACGGATGTT) IR2: 115..122, 130..137 (AAGTCGTT..AACGACTT) IR3: 141..146, 151..156 (CGCACC..GGTGCG) IR4: 230..236, 241..247 ( TGACCCT..AGGGTCA) IR5: 256..265, 269..278 (CCCAATGCGC..GCGCATTGGG) |
| Location of nic site | 164..165 |
| Conserved sequence flanking the nic site |
TGTCT|ATAGC |
| Note | The oriT of pHH2-227 is almost identical to that of R7K. |
oriT sequence
Download Length: 341 nt
GTGCCATCTGCGCTTTGTGGTCCTAGTTGTGGTCCCAAAAGTATAGCAAACTCGGGCTATTACGCAACATCTGTGGTCCCTGTAGACCACGGATGTTTAGCGCCATTGCGCCGGAAGTCGTTGAAAATCAACGACTTACGCGCACCGAAAGGTGCGTATTGTCTATAGCCCAGATTTAAGGATACCAACCCTGCTTTTAAGGACGAAAACCATGCCGATAACGCCAGCGTGACCCTAAAGAGGGTCAAAACTGCTCCCAATGCGCTATGCGCATTGGGTTATCGTGCAACAATAATGCAACTATAATGCTATGATGGTGCGACAATGATGCAGAAAATCAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file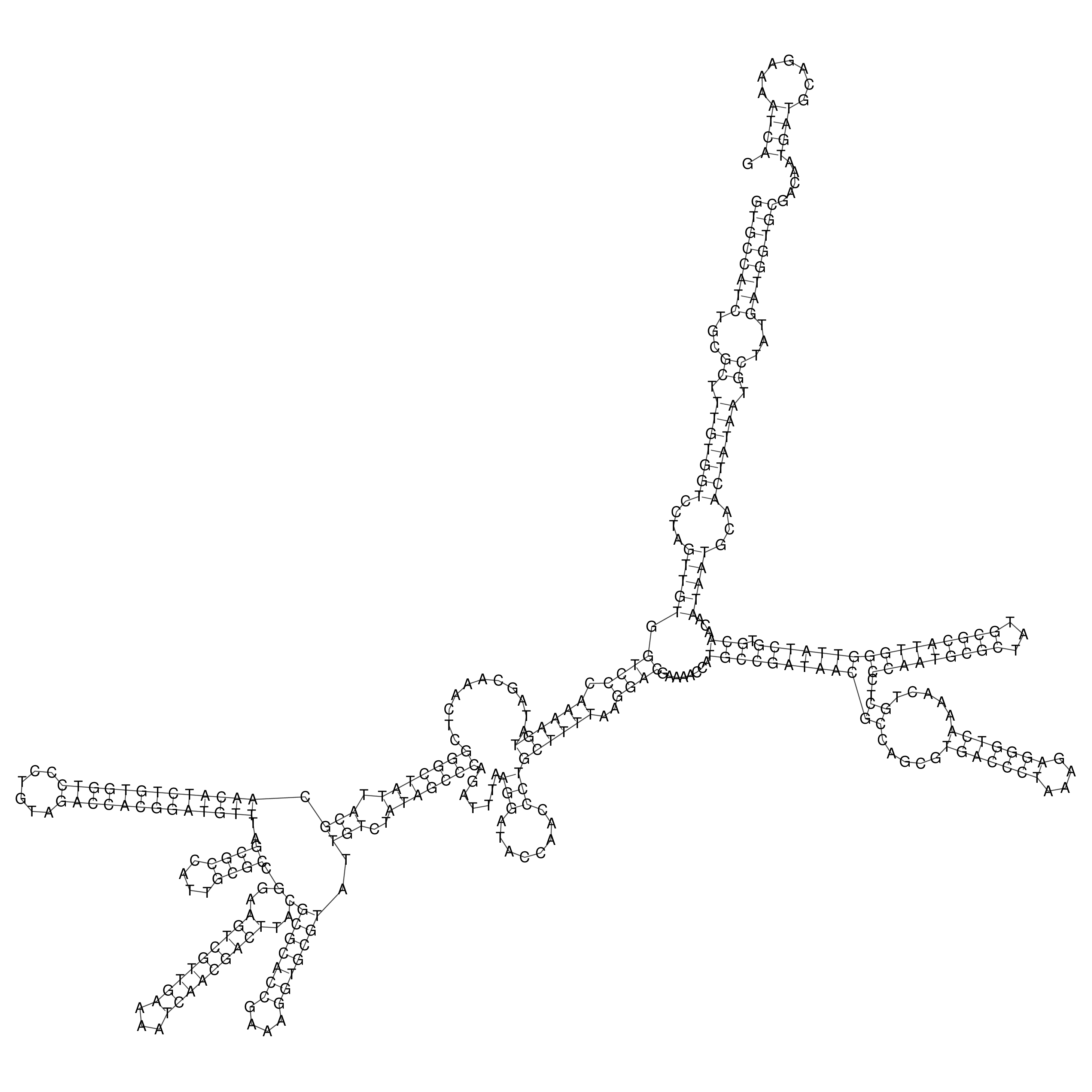
Reference
[1] Król JE et al. (2013) Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid. 70(1):110-9. [PMID:23558148]
Relaxase
| ID | 92 | GenBank | AEY63615 |
| Name | TrwC_pHH2-227 |
UniProt ID | H2DDW0 |
| Length | 966 a.a. | PDB ID | |
| Note | conjugative relaxase | ||
Relaxase protein sequence
Download Length: 966 a.a. Molecular weight: 107460.80 Da Isoelectric Point: 9.8804
MLSHMVLTRQDIGRAASYYEDGADDYYAKDGDASEWQGKGAEELGLSGEVDSKRFRELLAGNIGEGHRIM
RSATRQDSKERIGLDLTFSAPKSVSLQALVAGDAEIIKAHDRAVARTLEQAEARAQARQKIQGKTRIETT
GNLVIGKFRHETSRERDPQLHTHAVILNMTKRSDGQWRALKNDEIVKATRYLGAVYNAELAHELQKLGYQ
LRYGKDGNFDLAHIDRQQIEGFSKRTEQIAEWYAARGLDPNSVSLEQKQAAKVLSRAKKTSVDREALRAE
WQATAKELGIDFSRREWSGREKGGSEKQAHSFMPSDEAAKRAVRYAINHLTERQSVMDERELVDTAMKHA
VGAARLEDIQKELLRQTETGYLIREAPRYRPGGQTGPTDEPGKTRAEWIAELAAKGMKQGAARERVDNAI
KTGGLVPIEPRYTTQTALEREKRILQIERDGRGAVAPVIAAEAARERLASTNLNQGQREAAELIVSAANR
VVGVQGFAGTGKSHMLDTAKQMIEGEGYHVRALAPYGSQVKALRELNVEANTLASFLRAKDKNIDSRTVL
VIDEAGVVPTRLMEQTLKLAEKAGARVVLMGDTAQTKAIEAGRPFDQLQAAGMQTAHMREIQRQKNPELK
IAVELAAAGKASSSLERIKDVTEIKNHHERRAAVAEAYIALKPDERDRTLIVSGTNEARREINQIVREGL
GTAGKGIEFDTLVRVDTTQAERRHSKNYQVGHVIQPERDYAKTGLQRGELYRVVETGPGNRLTVIGEHDG
QRIQFSPMTHTKISVYQPERAELAVGDTIRITRNDKHLDLANGDRMKVVAVEDRKVTVTDGKRNVELPTD
KPLHVDHAYATTVHSSQGLTSDRVLIDAHAESRTTAKDVYYVAISRARFEARVFTNDRGKLPAAIARENI
KSAAHDLARDRGGRSAAAERQREQQREAERNRQTQQPAHDRQKAAREAERGMEAGR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | H2DDW0 |
Reference
[1] Król JE et al. (2013) Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid. 70(1):110-9. [PMID:23558148]
Auxiliary protein
| ID | 133 | GenBank | AEY63617 |
| Name | TrwA_pHH2-227 |
UniProt ID | H2DDW2 |
| Length | 121 a.a. | PDB ID | _ |
| Note | relaxosome accessory protein TrwA | ||
Auxiliary protein sequence
Download Length: 121 a.a. Molecular weight: 13473.38 Da Isoelectric Point: 5.0435
MALGEPIKFRLTPEKHAQYEDEAARLGKPLGTYLRERLEADDAVRDELAALRREVVSLHHVIEDLADTGL
RSDQSGPGPNAVQIETLLLLRAIAGPERMKPVKGELKRLGIEVWTPEGKED
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | H2DDW2 |
T4CP
| ID | 92 | GenBank | AEY63616 |
| Name | TrwB_pHH2-227 |
UniProt ID | H2DDW1 |
| Length | 507 a.a. | PDB ID | _ |
| Note | coupling protein TrwB | ||
T4CP protein sequence
Download Length: 507 a.a. Molecular weight: 56258.97 Da Isoelectric Point: 9.9420
MHPDDQRKVSAGIVIVLPLIFWITAVQKTEVLGSPKLLALWELMKLTPQKPILLLSALGGLAVGVLFVWL
LNSVGQGEFGGAPFKRFLRGTRIVSGGKLKRMTREKAKQVTVAGVPMPRDAEPRHLLVNGATGTGKSVLL
RELAYTGLLRGDRMVIVDPNGDMLSKFGRDKDVILNPYDKRTKGWSFFNEIRNDYDWQRYALSVVPRGKT
DEAEEWASYGRLLLRETAKKLALIGTPSMRELFHWTTIATFDDLRGFLEGTLAESLFAGSNEASKALTSA
RFVLSDKLPEHVTMPDGDFSIRSWLEDPNGGNLFITWREDMGPALRPLISAWVDVVCTSILSLPEEPKRR
LWLFIDELASLEKLASLADALTKGRKAGLRVVAGLQSTSQLDDVYGVKEAQTLRASFRSLVVLGGSRTDP
KTNEDMSLSLGEHEVERDRYSKNTGKHHSTGRALERVRERVVMPAEIANLPDLTAYVGFAGNRPIAKVPL
EIKQFANRQPAFVEGTI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | H2DDW1 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 7629..22267
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_3 | 3719..4732 | - | 1014 | AEY63597 | site specific recombinase | - |
| Locus_4 | 4849..5685 | + | 837 | AEY63598 | dihydropteroate synthase | - |
| Locus_5 | 5813..6313 | + | 501 | AEY63599 | acetyltransferase, gnat family | - |
| Locus_6 | 6610..6897 | - | 288 | AEY63600 | KorB | - |
| Locus_7 | 7034..7627 | + | 594 | AEY63601 | exported protein KikA | - |
| Locus_8 | 7629..8327 | + | 699 | AEY63602 | conjugal transfer protein | virB1 |
| Locus_9 | 8518..8745 | + | 228 | AEY63603 | putative transcriptional repressor KorA | - |
| Locus_10 | 8711..9016 | + | 306 | AEY63604 | conjugal transfer protein | virB2 |
| Locus_11 | 9029..9343 | + | 315 | AEY63605 | conserved hypothetical plasmid protein | virB3 |
| Locus_12 | 9346..11817 | + | 2472 | AEY63606 | conjugal transfer protein | virb4 |
| Locus_13 | 11814..12503 | + | 690 | AEY63607 | conjugal transfer protein | virB5 |
| Locus_14 | 12500..12730 | + | 231 | AEY63608 | entry exclusion protein | - |
| Locus_15 | 12744..13772 | + | 1029 | AEY63609 | conjugal transfer protein | virB6 |
| Locus_16 | 13885..14028 | + | 144 | AEY63610 | conjugal transfer protein | - |
| Locus_17 | 14025..14720 | + | 696 | AEY63611 | conjugal transfer protein | virB8 |
| Locus_18 | 14732..15532 | + | 801 | AEY63612 | conjugal transfer protein | virB9 |
| Locus_19 | 15532..16719 | + | 1188 | AEY63613 | conjugal transfer protein | virB10 |
| Locus_20 | 16748..17761 | + | 1014 | AEY63614 | conjugal transfer protein | virB11 |
| Locus_21 | 17844..20744 | - | 2901 | AEY63615 | conjugative relaxase | - |
| Locus_22 | 20744..22267 | - | 1524 | AEY63616 | coupling protein TrwB | virb4 |
| Locus_23 | 22269..22634 | - | 366 | AEY63617 | relaxosome accessory protein TrwA | - |
| Locus_24 | 23090..23515 | + | 426 | AEY63618 | conserved hypothetical plasmid protein | - |
| Locus_25 | 23512..24207 | + | 696 | AEY63619 | conserved hypothetical plasmid protein | - |
| Locus_26 | 24221..24607 | + | 387 | AEY63620 | plasmid membrane protein | - |
| Locus_27 | 24763..25014 | - | 252 | AEY63621 | conserved hypothetical plasmid protein | - |
| Locus_28 | 25162..25815 | - | 654 | AEY63622 | conserved hypothetical plasmid protein | - |
| Locus_29 | 25831..26103 | - | 273 | AEY63623 | conserved hypothetical plasmid protein | - |
| Locus_30 | 26868..27191 | - | 324 | AEY63624 | transcriptional regulator | - |
Host bacterium
| ID | 92 | GenBank | JN581942 |
| Plasmid name | pHH2-227 | Incompatibility group | IncW |
| Plasmid size | 38963 bp | Coordinate of oriT [Strand] | |
| Host baterium | Uncultured bacterium |
Cargo genes
| Drug resistance gene | resistance to tetracycline and sulfonamide |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |