Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100094 |
| Name | oriT_pNDM-40-1 |
| Organism | Acinetobacter bereziniae strain CHI-40-1 |
| Sequence Completeness | incomplete |
| NCBI accession of oriT (coordinates [strand]) | KF702385 (1..38 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 8..17, 21..30 (CATAAGGGAA..TTCCCTTATG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note |
oriT sequence
Download Length: 38 nt
>oriT_pNDM-40-1
AGGGATTCATAAGGGAATTATTCCCTTATGTGGGGCTT
AGGGATTCATAAGGGAATTATTCCCTTATGTGGGGCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file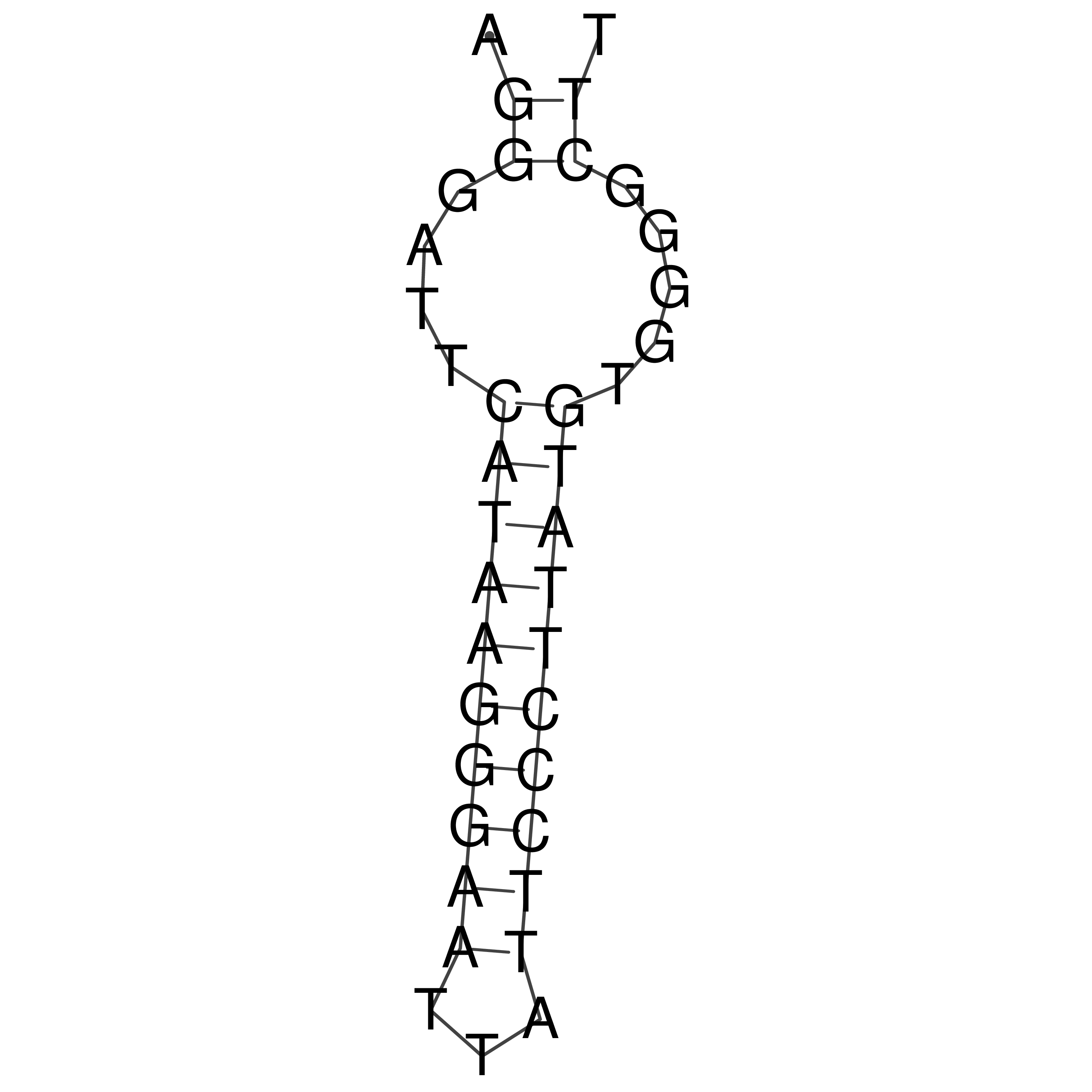
Reference
[1] Jones LS et al. (2015) Characterization of plasmids in extensively drug-resistant acinetobacter strains isolated in India and Pakistan. Antimicrob Agents Chemother. 59(2):923-9. [PMID:25421466]
Relaxase
| ID | 89 | GenBank | AHF22521 |
| Name | TraA_pNDM-40-1 |
UniProt ID | W0FF73 |
| Length | 996 a.a. | PDB ID | |
| Note | conjugal transfer relaxase | ||
Relaxase protein sequence
Download Length: 996 a.a. Molecular weight: 113758.44 Da Isoelectric Point: 9.0108
>AHF22521.1 TraA (plasmid) [Acinetobacter bereziniae]
MTIAVGMHASAFLSEKQKKQKQKRHKMAIYHLSVKTGRQYGSTGTRGANHYAYINRDGKYKRDDLENIKS
GNLPSWATDAKHFWKSADKNERINGRVYTELEISLPRELNKEQREELVEKFVEKTLGENFTYSYAIHSPL
ASDGQQNPHVHLMFSERKLDGIERDEVKHFKRYNPVNPEKGGAGKDRYFSSKMFVVDTRLEWANHVNDFC
ENLGIDARIDHRSFKEQGIELSSQNFRADYVSSHKYHINENIKQVRHQNGEIIIERPSEAIKALTSNQSV
FSVRELERFLMAHTDGEEQYLKAYESVITCPDMATLYGKDKVVFSSKELVGIEESISTFIYNANEESRIQ
KPFNLVEKLPLNAVRVAETRTFNSEQEKAFYTLTSTDRITLLNGSAGTGKSYVLSAVSEAYKESDYQVYG
IALQAITAKAIANDCDIPSSTIASFLSKYDSGNLEINNKTVLILDEAGMVGSRDMQRLLMLVEKHDAQIK
LVGDSYQLSAVSAGHVFANIQENLDKKNIASLEDIQRQKHAKESDKMIQASKYLSKHEVGKAIDIYKSIN
MVNEYESHDLALAKTVKSWSEDSSESKLMLAHSNSDVNELNRMARALLIKQGKLNPESVSVKNYQGDLDI
SIGEKIVFKQNNSQMKVSNGELAKVIGFEKDKFNNPKAMMVQMESDSRIVRVDLKEYDQFKHGYANTIHS
SQGMTVDSAYIVASENMNANLTYVALTRHKQNIQINYSALTFATNEKETVKTPDGKVSYDHNYKPVEREV
TAFENFKKVLGRSETKEFSTDFTLVESKDNLMKSYLNDHRLHLENKRELFGINSKLYSLTSENKTFTKEE
IVSEVRASLKSKALLGDEIVKFNGNMNIAKGELVKLTNDLEIKTGLFKKVTFEKGTELKVIDAYKVKDKP
VMKARIKNEEYEIPFNKLNLEYSDKYFNEFKDQSKARFDSRHKTEVYTEFREQLLKQKANAPKDNLNAPN
QRIKNPNTPNRNTPKY
MTIAVGMHASAFLSEKQKKQKQKRHKMAIYHLSVKTGRQYGSTGTRGANHYAYINRDGKYKRDDLENIKS
GNLPSWATDAKHFWKSADKNERINGRVYTELEISLPRELNKEQREELVEKFVEKTLGENFTYSYAIHSPL
ASDGQQNPHVHLMFSERKLDGIERDEVKHFKRYNPVNPEKGGAGKDRYFSSKMFVVDTRLEWANHVNDFC
ENLGIDARIDHRSFKEQGIELSSQNFRADYVSSHKYHINENIKQVRHQNGEIIIERPSEAIKALTSNQSV
FSVRELERFLMAHTDGEEQYLKAYESVITCPDMATLYGKDKVVFSSKELVGIEESISTFIYNANEESRIQ
KPFNLVEKLPLNAVRVAETRTFNSEQEKAFYTLTSTDRITLLNGSAGTGKSYVLSAVSEAYKESDYQVYG
IALQAITAKAIANDCDIPSSTIASFLSKYDSGNLEINNKTVLILDEAGMVGSRDMQRLLMLVEKHDAQIK
LVGDSYQLSAVSAGHVFANIQENLDKKNIASLEDIQRQKHAKESDKMIQASKYLSKHEVGKAIDIYKSIN
MVNEYESHDLALAKTVKSWSEDSSESKLMLAHSNSDVNELNRMARALLIKQGKLNPESVSVKNYQGDLDI
SIGEKIVFKQNNSQMKVSNGELAKVIGFEKDKFNNPKAMMVQMESDSRIVRVDLKEYDQFKHGYANTIHS
SQGMTVDSAYIVASENMNANLTYVALTRHKQNIQINYSALTFATNEKETVKTPDGKVSYDHNYKPVEREV
TAFENFKKVLGRSETKEFSTDFTLVESKDNLMKSYLNDHRLHLENKRELFGINSKLYSLTSENKTFTKEE
IVSEVRASLKSKALLGDEIVKFNGNMNIAKGELVKLTNDLEIKTGLFKKVTFEKGTELKVIDAYKVKDKP
VMKARIKNEEYEIPFNKLNLEYSDKYFNEFKDQSKARFDSRHKTEVYTEFREQLLKQKANAPKDNLNAPN
QRIKNPNTPNRNTPKY
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W0FF73 |
Reference
[1] Jones LS et al. (2015) Characterization of plasmids in extensively drug-resistant acinetobacter strains isolated in India and Pakistan. Antimicrob Agents Chemother. 59(2):923-9. [PMID:25421466]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21010..34305
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_15 | 16888..17535 | - | 648 | AHF22506 | resolvase | - |
| Locus_16 | 18095..19807 | + | 1713 | AHF22505 | putative zeta toxin family protein | - |
| Locus_17 | 19804..20286 | - | 483 | AHF22504 | putative nuclease | - |
| Locus_18 | 20486..20947 | - | 462 | AHF22503 | hypothetical_protein | - |
| Locus_19 | 21010..21651 | - | 642 | AHF22502 | VirB5 | virB5 |
| Locus_20 | 21648..24359 | - | 2712 | AHF22501 | VirB4 | virb4 |
| Locus_21 | 24371..24664 | - | 294 | AHF22500 | VirB2 | virB2 |
| Locus_22 | 24675..25391 | - | 717 | AHF22499 | VirB1 | - |
| Locus_23 | 25430..25630 | - | 201 | AHF22498 | hypothetical protein | - |
| Locus_24 | 25649..27166 | - | 1518 | AHF22497 | VirD4 | virb4 |
| Locus_25 | 27163..28416 | - | 1254 | AHF22496 | putative transglycosylase | - |
| Locus_26 | 28492..28944 | - | 453 | AHF22495 | hypothetical protein | - |
| Locus_27 | 28948..29322 | - | 375 | AHF22494 | hypothetical protein | - |
| Locus_28 | 29246..30283 | - | 1038 | AHF22493 | VirB11 | virB11 |
| Locus_29 | 30299..31678 | - | 1380 | AHF22492 | VirB10 | virB10 |
| Locus_30 | 31680..32417 | - | 738 | AHF22491 | VirB9 | virB9 |
| Locus_31 | 32432..33220 | - | 789 | AHF22490 | VirB8 | virB8 |
| Locus_32 | 33220..34305 | - | 1086 | AHF22489 | VirB6 | virB6 |
| Locus_33 | 34313..34750 | - | 438 | AHF22488 | hypothetical protein | - |
| Locus_34 | 34719..35090 | - | 372 | AHF22487 | hypothetical protein | - |
| Locus_35 | 35087..35287 | - | 201 | AHF22486 | hypothetical protein | - |
| Locus_36 | 36712..36951 | + | 240 | AHF22485 | hypothetical protein | - |
| Locus_37 | 36968..37657 | + | 690 | AHF22484 | ParA | - |
| Locus_38 | 37654..38058 | + | 405 | AHF22483 | hypothetical protein | - |
| Locus_39 | 38068..38256 | + | 189 | AHF22482 | hypothetical protein | - |
Host bacterium
| ID | 89 | GenBank | KF702385 |
| Plasmid name | pNDM-40-1 | Incompatibility group | - |
| Plasmid size | 45826 bp | Coordinate of oriT [Strand] | 1..38 [+] |
| Host baterium | Acinetobacter bereziniae strain CHI-40-1 |
Cargo genes
| Drug resistance gene | harboring blaNDM-1 gene (encoding \New Delhi metal-beta-lactamase enzyme\) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |