Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100058 |
| Name | oriT2_pSBardo-Kan |
| Organism | Salmonella enterica subsp. enterica serovar Bardo |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_015575 ( 1522..1790 [-], 269 nt) |
| oriT length | 269 nt |
| IRs (inverted repeats) | 223..230, 233..240 (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 249..250 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | oriT; F-type |
oriT sequence
Download Length: 269 nt
>oriT2_pSBardo-Kan
CACCCCTAGCAGCGCCCCCAGGGGTGTCCTGTAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAGATGATGTTTTTTATGAAAATAGTCAGTACCACCCCTAAAAAGCGGTGTTGGCGCAGCTACAACACCGCGCCGACACCGCTTTTTTAAATATCACAAAGAGAGCAAGAAGAACTAATTTGTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGGCATTTCAGGCATGAGAA
CACCCCTAGCAGCGCCCCCAGGGGTGTCCTGTAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAGATGATGTTTTTTATGAAAATAGTCAGTACCACCCCTAAAAAGCGGTGTTGGCGCAGCTACAACACCGCGCCGACACCGCTTTTTTAAATATCACAAAGAGAGCAAGAAGAACTAATTTGTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGGCATTTCAGGCATGAGAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file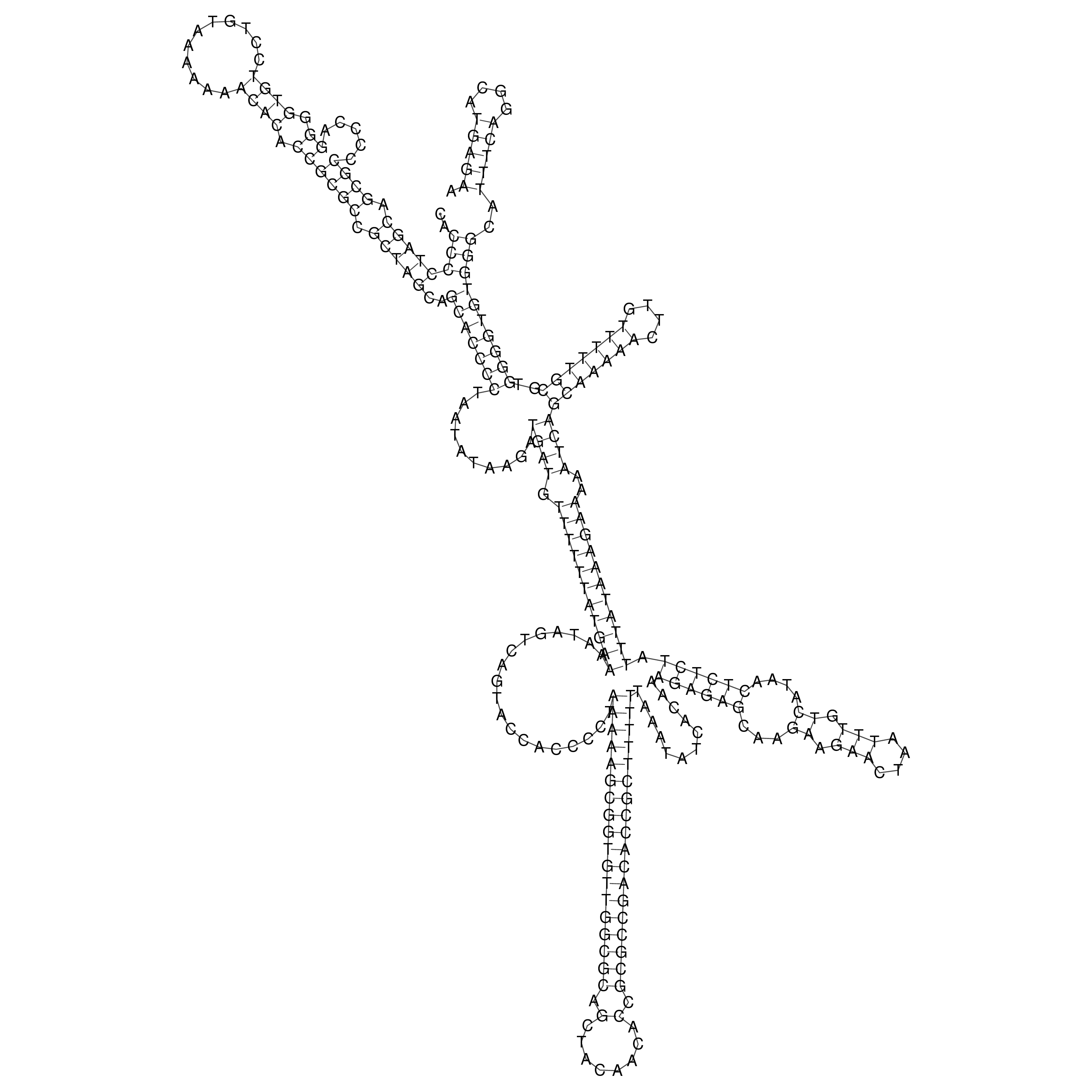
Reference
[1] Chen CY et al. (2011) Sequence analysis of a group of low molecular-weight plasmids carrying multiple IS903 elements flanking a kanamycin resistance aph gene in Salmonella enterica serovars. Plasmid. 65(3):246-52. [PMID:21324339]
Host bacterium
| ID | 55 | GenBank | NC_015575 |
| Plasmid name | pSBardo-Kan | Incompatibility group | ColRNAI |
| Plasmid size | 8198 bp | Coordinate of oriT [Strand] | 936..993 [-]; 1522..1790 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Bardo |
Cargo genes
| Drug resistance gene | aph(3')-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |