Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100033 |
| Name | oriT_pSTM709 |
| Organism | Salmonella enterica subsp. enterica serovar Typhimurium |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_023915 (_) |
| oriT length | 88 nt |
| IRs (inverted repeats) | 8..bp: 2..9, 13..20 (GTCGGGGC..GCCCTGAC) 17..bp: 27..43, 50..66 (GTAATTGTAATAGCGTC..GACGGTATTACAATTAC) |
| Location of nic site | 74..75 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | _ |
oriT sequence
Download Length: 88 nt
>oriT_pSTM709
TGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
TGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file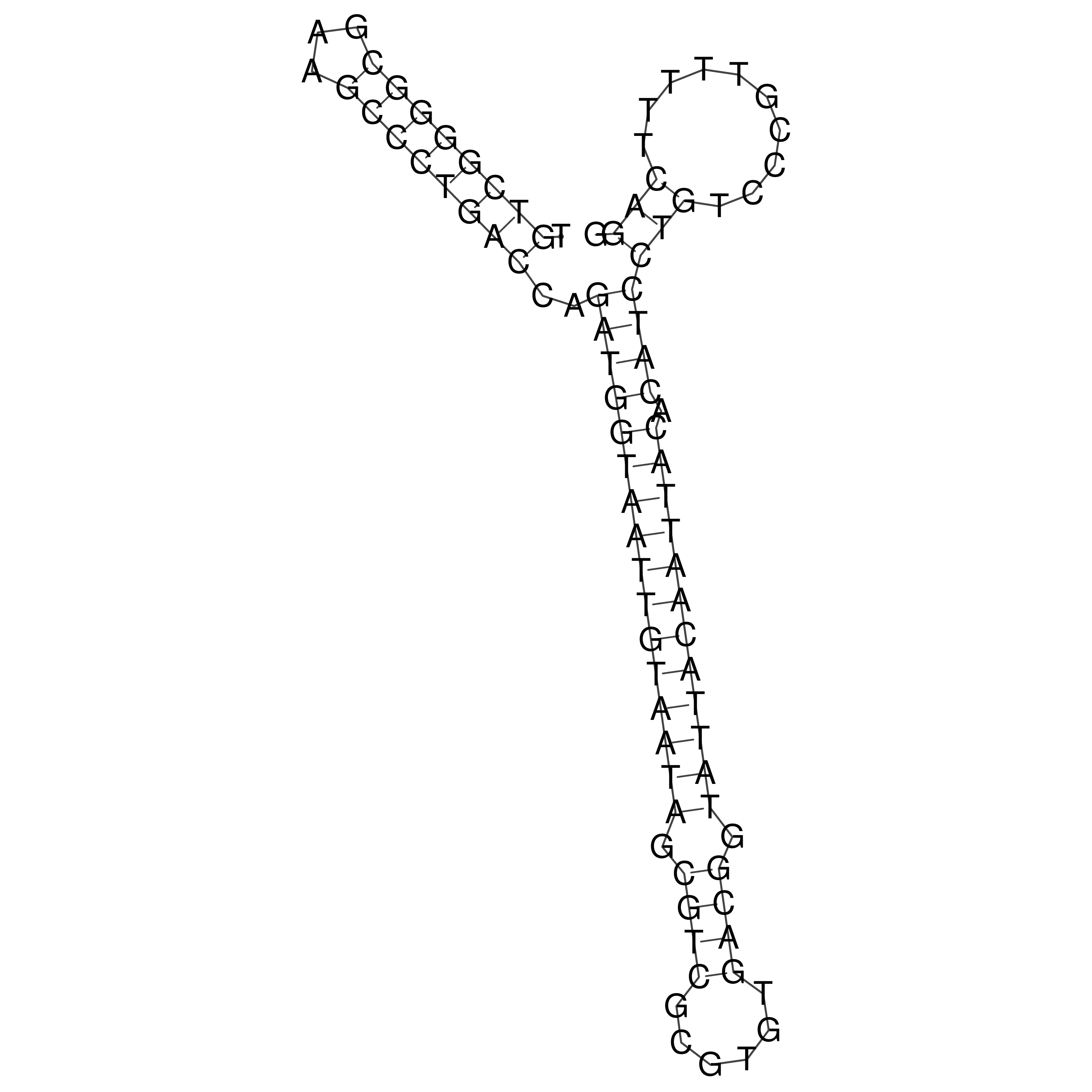
Relaxase
| ID | 32 | GenBank | YP_009023753 |
| Name | NikB_pSTM709 |
UniProt ID | X5JA84 |
| Length | 899 a.a. | PDB ID | |
| Note | NikB relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103834.33 Da Isoelectric Point: 7.3473
>YP_009023753.1 NikB relaxase (plasmid) [Salmonella enterica subsp. enterica serovar Typhimurium str. STM709]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 49 | GenBank | YP_009023752 |
| Name | NikA_pSTM709 |
UniProt ID | X5J9V3 |
| Length | 110 a.a. | PDB ID | _ |
| Note | NikA oriT-specific DNA binding protein | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
>YP_009023752.1 NikA oriT-specific DNA binding protein (plasmid) [Salmonella enterica subsp. enterica serovar Typhimurium str. STM709]
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 32 | GenBank | YP_009023754 |
| Name | TrbC_pSTM709 |
UniProt ID | X5JAP4 |
| Length | 763 a.a. | PDB ID | _ |
| Note | TrbC transfer protein | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86927.12 Da Isoelectric Point: 6.7713
>YP_009023754.1 TrbC transfer protein (plasmid) [Salmonella enterica subsp. enterica serovar Typhimurium str. STM709]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKIDVQELIELH
PGEFFSIFRGETVPSASFFIPDNEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTAQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKIDVQELIELH
PGEFFSIFRGETVPSASFFIPDNEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTAQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 49375..89077
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| D645_p16051 | 47091..49382 | - | 2292 | YP_009023754 | TrbC transfer protein | - |
| D645_p16050 | 49375..50445 | - | 1071 | YP_009023755 | IncI1 conjugal transfer protein TrbB | trbB |
| D645_p16049 | 50464..51672 | - | 1209 | YP_009023756 | TrbA transfer protein | trbA |
| D645_p16048 | 51906..52109 | + | 204 | YP_009023757 | post-segregation killing protein | - |
| D645_p16047 | 51964..52116 | + | 153 | YP_009023758 | post-segregation killing protein PndA | - |
| D645_p16046 | 52800..53582 | - | 783 | YP_009023759 | insertion sequence IS100, ATP-binding protein | - |
| D645_p16045 | 53579..54601 | - | 1023 | YP_009023760 | IS100 transposase orfA | - |
| D645_p16044 | 56171..56833 | - | 663 | YP_009023761 | surface exclusion protein ExcA | - |
| D645_p16043 | 56904..59141 | - | 2238 | YP_009023762 | TraY integral membrane protein | traY |
| D645_p16042 | 59169..59753 | - | 585 | YP_009023763 | conjugative transfer protein TraX | - |
| D645_p16040 | 60950..61564 | - | 615 | YP_009023764 | conjugative transfer protein TraV | traV |
| D645_p16039 | 61564..63591 | - | 2028 | YP_009023765 | TraU nucleotide-binding protein | traU |
| D645_p16038 | 64697..65497 | - | 801 | YP_009023766 | conjugative transfer protein TraT | traT |
| D645_p16037 | 65481..65669 | - | 189 | YP_009023767 | conjugal transfer protein TraS | - |
| D645_p16036 | 65733..66137 | - | 405 | YP_009023768 | conjugal transfer protein TraR | traR |
| D645_p16035 | 66188..66715 | - | 528 | YP_009023769 | conjugal transfer protein TraQ | traQ |
| D645_p16034 | 66715..67419 | - | 705 | YP_009023770 | conjugal transfer protein TraP | traP |
| D645_p16033 | 67419..68708 | - | 1290 | YP_009023771 | TraO transfer protein | traO |
| D645_p16032 | 68711..69694 | - | 984 | YP_009023772 | conjugative transfer protein TraN | traN |
| D645_p16031 | 69705..70397 | - | 693 | YP_009023773 | conjugal transfer protein TraM | traM |
| D645_p16030 | 70394..70741 | - | 348 | YP_009023774 | conjugal transfer protein TraL | traL |
| D645_p16028 | 70759..74562 | - | 3804 | YP_009023775 | SogL DNA primase | - |
| D645_p16027 | 74616..75167 | - | 552 | YP_009023776 | EDTA-resistant nuclease | - |
| D645_p16026 | 75182..75472 | - | 291 | YP_009023777 | conjugal protein Trak | traK |
| D645_p16025 | 75469..76617 | - | 1149 | YP_009023778 | plasmid transfer ATPase TraJ | virB11 |
| D645_p16024 | 76614..77432 | - | 819 | YP_009023779 | conjugal transfer protein TraI | traI |
| D645_p16023 | 77429..77887 | - | 459 | YP_009023780 | conjugal transfer protein TraH | - |
| D645_p16022 | 78282..78866 | - | 585 | YP_009023781 | conjugal transfer protein TraG | - |
| D645_p16021 | 78926..80128 | - | 1203 | YP_009023782 | conjugal transfer protein TraF | - |
| D645_p16020 | 80214..81038 | - | 825 | YP_009023783 | conjugal transfer protein TraE | traE |
| D645_p16019 | 81189..82343 | - | 1155 | YP_009023784 | Shufflon-specific DNA recombinase | - |
| D645_p16018 | 83147..84583 | - | 1437 | YP_009023785 | type III prepilin | - |
| D645_p16017 | 84571..85227 | - | 657 | YP_009023786 | Type-IV sectretion leader peptidase/N-methyltransferase PilU | - |
| D645_p16016 | 85212..85772 | - | 561 | YP_009023787 | putative transglycosylase PilT | virB1 |
| D645_p16015 | 85782..86396 | - | 615 | YP_009023788 | type IV prepilin PilS | - |
| D645_p16014 | 86414..87511 | - | 1098 | YP_009023789 | integral membrane protein PilR | - |
| D645_p16013 | 87524..89077 | - | 1554 | YP_009023790 | ATP-binding protein PilQ | virB11 |
| D645_p16012 | 89088..89540 | - | 453 | YP_009023791 | type IV pilus biogenesis protein PilP | - |
| D645_p16011 | 89527..90822 | - | 1296 | YP_009023792 | PilO type IV pilus biogenesis protein | - |
| D645_p16010 | 90815..92497 | - | 1683 | YP_009023793 | PilN type IV pilus secretin | - |
| D645_p16009 | 92511..92948 | - | 438 | YP_009023794 | conjugal transfer protein PilM | - |
| D645_p16008 | 92948..94015 | - | 1068 | YP_009023795 | lipoprotein PilL | - |
Host bacterium
| ID | 32 | GenBank | NC_023915 |
| Plasmid name | pSTM709 | Incompatibility group | IncI1 |
| Plasmid size | 99184 bp | Coordinate of oriT [Strand] | 43906..43949 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Typhimurium |
Cargo genes
| Drug resistance gene | class-C beta-lactamase gene blaCMY-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |