Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100028 |
| Name | oriT_R621a |
| Organism | Salmonella enterica subsp. enterica serovar Typhimurium |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_015965 (37332..37414 [-], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | 8..bp: 6..13, 17..24 (GTCGGGGC..GCCCTGAC) 17..bp: 31..754..70 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 78..79 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | minimal oriT sequence |
oriT sequence
Download Length: 83 nt
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file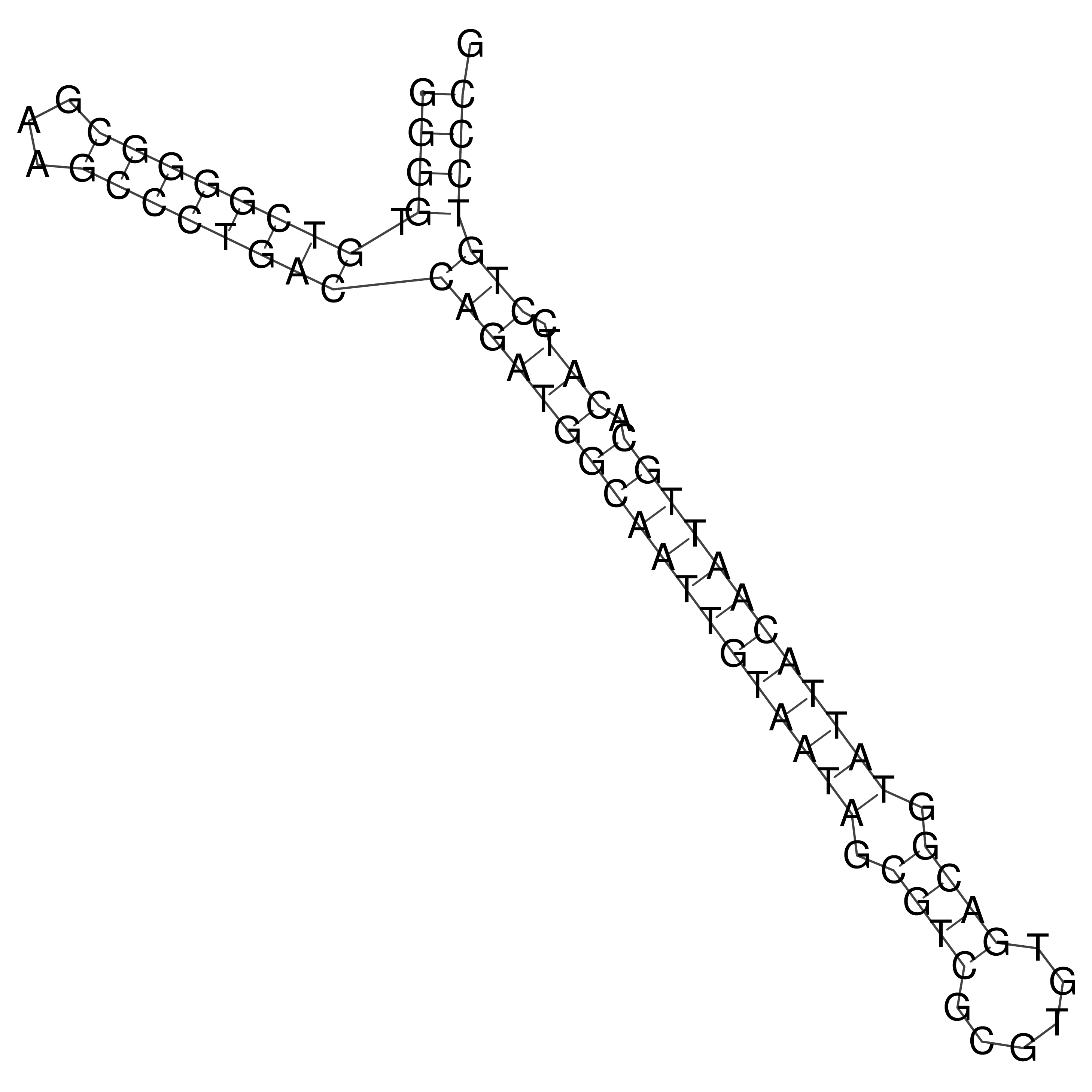
Reference
[1] Takahashi H et al. (2011) The genome sequence of the incompatibility group I纬 plasmid R621a: evolution of IncI plasmids. Plasmid. 66(2):112-21. [PMID:21763721]
Relaxase
| ID | 27 | GenBank | YP_004823733 |
| Name | NikB_R621a |
UniProt ID | G2HK77 |
| Length | 899 a.a. | PDB ID | |
| Note | NikB relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104019.46 Da Isoelectric Point: 7.3526
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | G2HK77 |
Reference
[1] Takahashi H et al. (2011) The genome sequence of the incompatibility group I纬 plasmid R621a: evolution of IncI plasmids. Plasmid. 66(2):112-21. [PMID:21763721]
Auxiliary protein
| ID | 41 | GenBank | YP_004823732 |
| Name | NikA_R621a |
UniProt ID | Q79VV8 |
| Length | 111 a.a. | PDB ID | _ |
| Note | NikA oriT-specific DNA-binding protein | ||
Auxiliary protein sequence
Download Length: 111 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q79VV8 |
Reference
[1] Takahashi H et al. (2011) The genome sequence of the incompatibility group I纬 plasmid R621a: evolution of IncI plasmids. Plasmid. 66(2):112-21. [PMID:21763721]
T4CP
| ID | 27 | GenBank | YP_004823734 |
| Name | TrbC_R621a |
UniProt ID | G2HK78 |
| Length | 763 a.a. | PDB ID | _ |
| Note | TrbC transfer protein | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86914.98 Da Isoelectric Point: 6.6402
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTMITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILTAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLNTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | G2HK78 |
Reference
[1] Takahashi H et al. (2011) The genome sequence of the incompatibility group I纬 plasmid R621a: evolution of IncI plasmids. Plasmid. 66(2):112-21. [PMID:21763721]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 42792..83793
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HXF67_RS00250 (R621a_p053) | 40508..42799 | - | 2292 | WP_014065723 | F-type conjugative transfer protein TrbC | - |
| HXF67_RS00255 (R621a_p054) | 42792..43862 | - | 1071 | WP_014065724 | IncI1-type conjugal transfer protein TrbB | trbB |
| HXF67_RS00260 (R621a_p055) | 43881..45089 | - | 1209 | WP_001703845 | IncI1-type conjugal transfer protein TrbA | trbA |
| HXF67_RS00265 (R621a_p056) | 45396..46175 | - | 780 | WP_275542888 | protein FinQ | - |
| HXF67_RS00270 (R621a_p058) | 46802..46954 | + | 153 | WP_001331364 | Hok/Gef family protein | - |
| HXF67_RS00275 | 47026..47277 | - | 252 | WP_001291963 | hypothetical protein | - |
| HXF67_RS00545 | 47778..47873 | + | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| HXF67_RS00285 | 47938..48114 | - | 177 | WP_001054900 | hypothetical protein | - |
| HXF67_RS00290 | 48481..48690 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| HXF67_RS00295 (R621a_p059) | 48762..49376 | - | 615 | WP_000578648 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HXF67_RS00300 (R621a_p060) | 49452..51620 | - | 2169 | WP_000698360 | IncI1-type conjugal transfer membrane protein TraY | traY |
| HXF67_RS00305 (R621a_p061) | 51717..52301 | - | 585 | WP_001037993 | IncI1-type conjugal transfer protein TraX | - |
| HXF67_RS00310 (R621a_p062) | 52330..53532 | - | 1203 | WP_014065727 | IncI1-type conjugal transfer protein TraW | traW |
| HXF67_RS00315 (R621a_p063) | 53499..54113 | - | 615 | WP_014065728 | IncI1-type conjugal transfer protein TraV | traV |
| HXF67_RS00320 (R621a_p064) | 54113..57157 | - | 3045 | WP_014065729 | IncI1-type conjugal transfer protein TraU | traU |
| HXF67_RS00330 (R621a_p067) | 58583..59383 | - | 801 | WP_001493813 | IncI1-type conjugal transfer protein TraT | traT |
| HXF67_RS00335 (R621a_p068) | 59367..59555 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| HXF67_RS00340 (R621a_p069) | 59619..60023 | - | 405 | WP_000086964 | IncI1-type conjugal transfer protein TraR | traR |
| HXF67_RS00345 (R621a_p070) | 60074..60601 | - | 528 | WP_001493812 | conjugal transfer protein TraQ | traQ |
| HXF67_RS00350 (R621a_p071) | 60601..61305 | - | 705 | WP_001493811 | IncI1-type conjugal transfer protein TraP | traP |
| HXF67_RS00355 (R621a_p072) | 61305..62594 | - | 1290 | WP_014065730 | conjugal transfer protein TraO | traO |
| HXF67_RS00360 (R621a_p073) | 62597..63580 | - | 984 | WP_001191880 | IncI1-type conjugal transfer protein TraN | traN |
| HXF67_RS00365 (R621a_p074) | 63591..64283 | - | 693 | WP_000138551 | DotI/IcmL family type IV secretion protein | traM |
| HXF67_RS00370 (R621a_p075) | 64280..64627 | - | 348 | WP_001493810 | hypothetical protein | traL |
| HXF67_RS00375 (R621a_p076) | 64645..68412 | - | 3768 | WP_014065731 | LPD7 domain-containing protein | - |
| HXF67_RS00380 (R621a_p078) | 68502..69053 | - | 552 | WP_012783942 | phospholipase D family protein | - |
| HXF67_RS00385 (R621a_p079) | 69068..69358 | - | 291 | WP_014065733 | hypothetical protein | traK |
| HXF67_RS00390 (R621a_p080) | 69355..70503 | - | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| HXF67_RS00395 (R621a_p081) | 70500..71318 | - | 819 | WP_014065734 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| HXF67_RS00400 (R621a_p082) | 71315..71773 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| HXF67_RS00405 (R621a_p083) | 72167..72751 | - | 585 | WP_000977522 | histidine phosphatase family protein | - |
| HXF67_RS00410 (R621a_p084) | 72811..74013 | - | 1203 | WP_014065735 | conjugal transfer protein TraF | - |
| HXF67_RS00415 (R621a_p085) | 74099..74923 | - | 825 | WP_014065736 | conjugal transfer protein TraE | traE |
| HXF67_RS00420 (R621a_p086) | 75074..76228 | - | 1155 | WP_001139955 | site-specific integrase | - |
| HXF67_RS00425 (R621a_p087) | 76212..76538 | + | 327 | WP_001213803 | hypothetical protein | - |
| HXF67_RS00430 (R621a_p089) | 76833..77090 | - | 258 | WP_001302706 | hypothetical protein | - |
| HXF67_RS00435 (R621a_p091) | 77403..77558 | - | 156 | WP_001302705 | hypothetical protein | - |
| HXF67_RS00550 | 77641..77865 | + | 225 | Protein_89 | hypothetical protein | - |
| HXF67_RS00445 (R621a_p093) | 77862..79286 | - | 1425 | WP_001389385 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HXF67_RS00450 (R621a_p095) | 79286..79942 | - | 657 | WP_001193553 | prepilin peptidase | - |
| HXF67_RS00455 (R621a_p096) | 79927..80487 | - | 561 | WP_000014116 | lytic transglycosylase domain-containing protein | virB1 |
| HXF67_RS00460 (R621a_p097) | 80497..81111 | - | 615 | WP_014065737 | type 4 pilus major pilin | - |
| HXF67_RS00465 (R621a_p098) | 81130..82227 | - | 1098 | WP_014065738 | type II secretion system F family protein | - |
| HXF67_RS00470 (R621a_p099) | 82240..83793 | - | 1554 | WP_014065739 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HXF67_RS00475 (R621a_p100) | 83804..84256 | - | 453 | WP_001247870 | type IV pilus biogenesis protein PilP | - |
| HXF67_RS00480 (R621a_p101) | 84243..85538 | - | 1296 | WP_014065740 | type 4b pilus protein PilO2 | - |
| HXF67_RS00485 (R621a_p102) | 85531..87213 | - | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| HXF67_RS00490 (R621a_p103) | 87227..87664 | - | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| HXF67_RS00495 (R621a_p104) | 87664..88731 | - | 1068 | WP_014065741 | type IV pilus biogenesis lipoprotein PilL | - |
Host bacterium
| ID | 27 | GenBank | NC_015965 |
| Plasmid name | R621a | Incompatibility group | IncI |
| Plasmid size | 93185 bp | Coordinate of oriT [Strand] | 37332..37414 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Typhimurium |
Cargo genes
| Drug resistance gene | tet(B) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrVA2 |