Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100021 |
| Name | oriT_pDN1 |
| Organism | Dichelobacter nodosus |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_002636 (804..892 [-], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | 5..10, 16..21 (GTTTCT..AGAAAC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TAAGTGCG|CCCT |
| Note | _ |
oriT sequence
Download Length: 89 nt
GGCAGTTTCTTGAAGAGAAACCGGTAAGTGCGCCCTCCCGCAAAATGACGGTCGGGGTTTGCGGTGTCTGTGCTCCGATGATAGCATAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file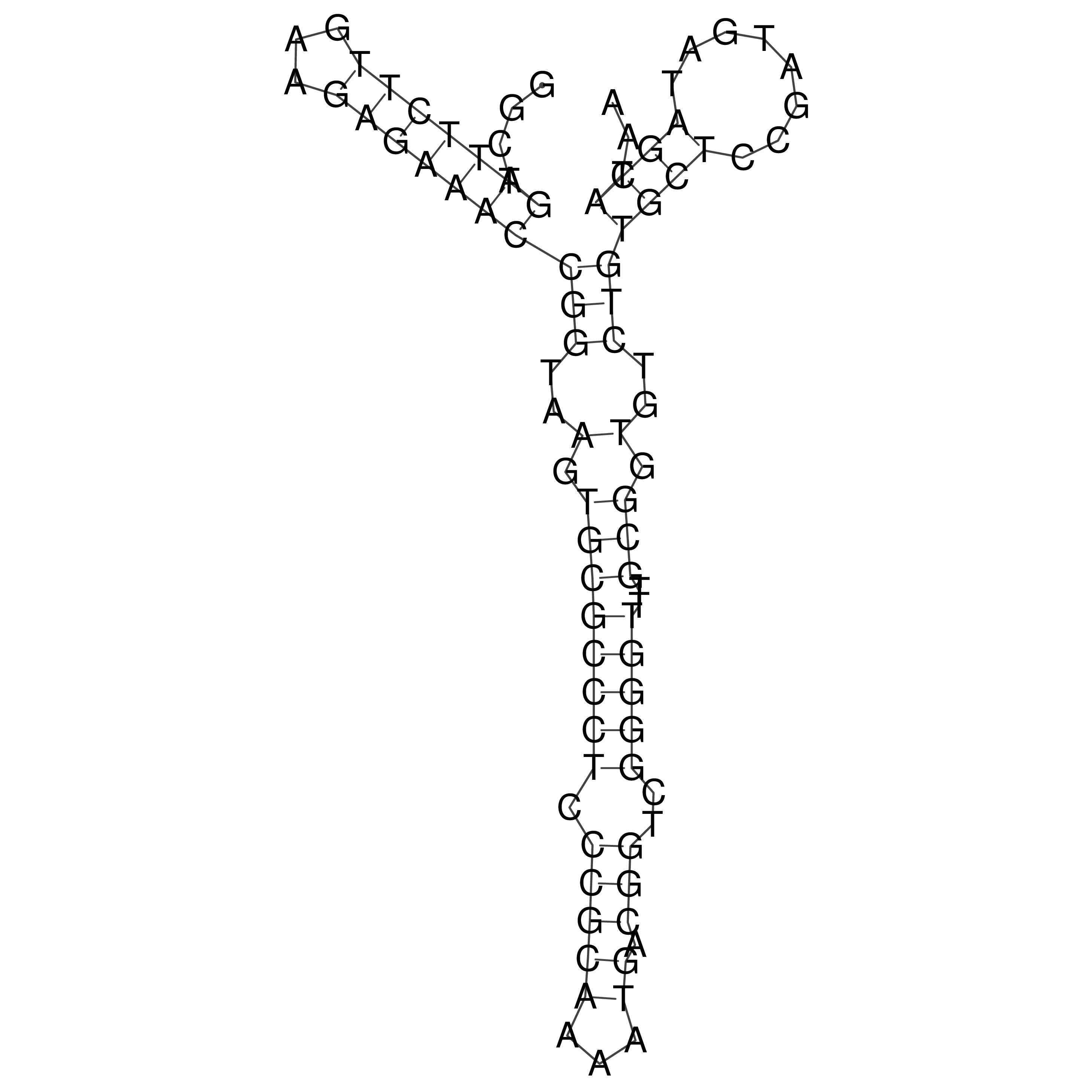
Reference
[1] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
[2] Whittle G et al. (2000) Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid. 43(3):230-4. [PMID:10783302]
Relaxase
| ID | 20 | GenBank | NP_073212 |
| Name | MobA_pDN1 |
UniProt ID | Q9EXP9 |
| Length | 703 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 703 a.a. Molecular weight: 78872.79 Da Isoelectric Point: 7.6998
MAIYHLTAKTGSRNGGQSAKAKADYIQREGRYSRDREEVLHTQSGHLPEWAERPADYWDGADLYERANGR
LFKEIEFALPVELTLDQQRELVDEFARHLTEGERLPYTLAIHAGNGENPHCHLMISERKNDGIERPADQW
FKRYNGKQPERGGAQKSESLKPKAWLEQTREAWSHYANRALEQAGHEARIDHRTLEAQGIERLPGIHLGP
NVVEMESRGIRTERADIALAIDTANGQIIDLQEYREVIEHERDRQSEEIQRDQRVSGRDRAAGPEHGDTG
GRSPAADRRGTAGQRGAGRELDEPATASHERMAAGSKGHEGSRRGTRQGRPDGPERRPWLDMEVVGRGLD
RFRDAYSGAADRIMALAGAANHRATGEHMADFQAQVKGDRTAQAIARQLKAMGCDRYDIGIRDAASGKMM
NREWTPQEVQQNAAWLKRMNAQGNDIYIRPAEQARHGLVLVDDLSSDDLDAMKQEGREPAAIIETSPKNY
QAWVKVAQDAPADHRGVIARKLAREYDADPASADSRHYGRLAGFTNRKDKYTSRTGYQPWVLCRESSGKS
ATAGPELMQQAGQVLDSIERRQERTARLAEITAPQSVRRYRRSVVDDYRSEMAGLVKRYGDDLSKCDFIA
AMKLASKGREADEIGKAMAEASPAIMERKAGHEADYIKRTVQKVMELPQVQEARAELAKQTQKQRSKGPD
LSM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9EXP9 |
Reference
[1] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
[2] Whittle G et al. (2000) Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid. 43(3):230-4. [PMID:10783302]
Auxiliary protein
| ID | 31 | GenBank | NP_073213 |
| Name | MobB_pDN1 |
UniProt ID | Q9EU19 |
| Length | 136 a.a. | PDB ID | _ |
| Note | _ | ||
Auxiliary protein sequence
Download Length: 136 a.a. Molecular weight: 15085.39 Da Isoelectric Point: 10.1732
MSAIDRVRKSRGINELAAEIEPLAQSMATLADEARQRIAEVQQASEEQAASWTSQQQQAMSAWRQAAKDM
RAAAGELAKAGQTARSAARGWTWRLWAGVLIASVMPILALLIASWLWLEPQIIEQQGSIWLIFKLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9EU19 |
Reference
[1] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
| ID | 32 | GenBank | NP_073211 |
| Name | MobC_pDN1 |
UniProt ID | Q9EXQ0 |
| Length | 99 a.a. | PDB ID | _ |
| Note | _ | ||
Auxiliary protein sequence
Download Length: 99 a.a. Molecular weight: 11617.14 Da Isoelectric Point: 10.3917
MAKTAERLAKLEEQRARINAEIQRIRAREQQQKRKENTRRKVLVGAWILGKVENGEWPEQRLLDGLDSYL
ERDHDRALFGLPPKQKQQPQEHGQPEPQG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q9EXQ0 |
Reference
[1] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
Host bacterium
| ID | 20 | GenBank | NC_002636 |
| Plasmid name | pDN1 | Incompatibility group | IncQ1 |
| Plasmid size | 5112 bp | Coordinate of oriT [Strand] | 804..892 [-] |
| Host baterium | Dichelobacter nodosus |
Cargo genes
| Drug resistance gene | _ |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |