Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100006 |
| Name | oriT_pIP501 |
| Organism | Streptococcus agalactiae B-49813/75 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | L39769 (1259..1296 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..12, 16..27 (ATACGAAGTAAC..GTTACTGCGTAT) |
| Location of nic site | 34..35 |
| Conserved sequence flanking the nic site |
TAAGTGC|GCCCT |
| Note | (1) The N-terminal domin (246 amino acids) of relaxase TraA_pIP501 show specific oriT binding [PMID:15554903]. (2) The relaxase TraA_pIP501 and its N-terminal domin (246 amino acids) behave as dimers in solution [PMID:15554903]. (3) The relaxase TraA_pIP501 negatively autoregulate the downstream 15 tra gene cluster [PMID:16514144]. |
oriT sequence
Download Length: 38 nt
ATACGAAGTAACGAAGTTACTGCGTATAAGTGCGCCCT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file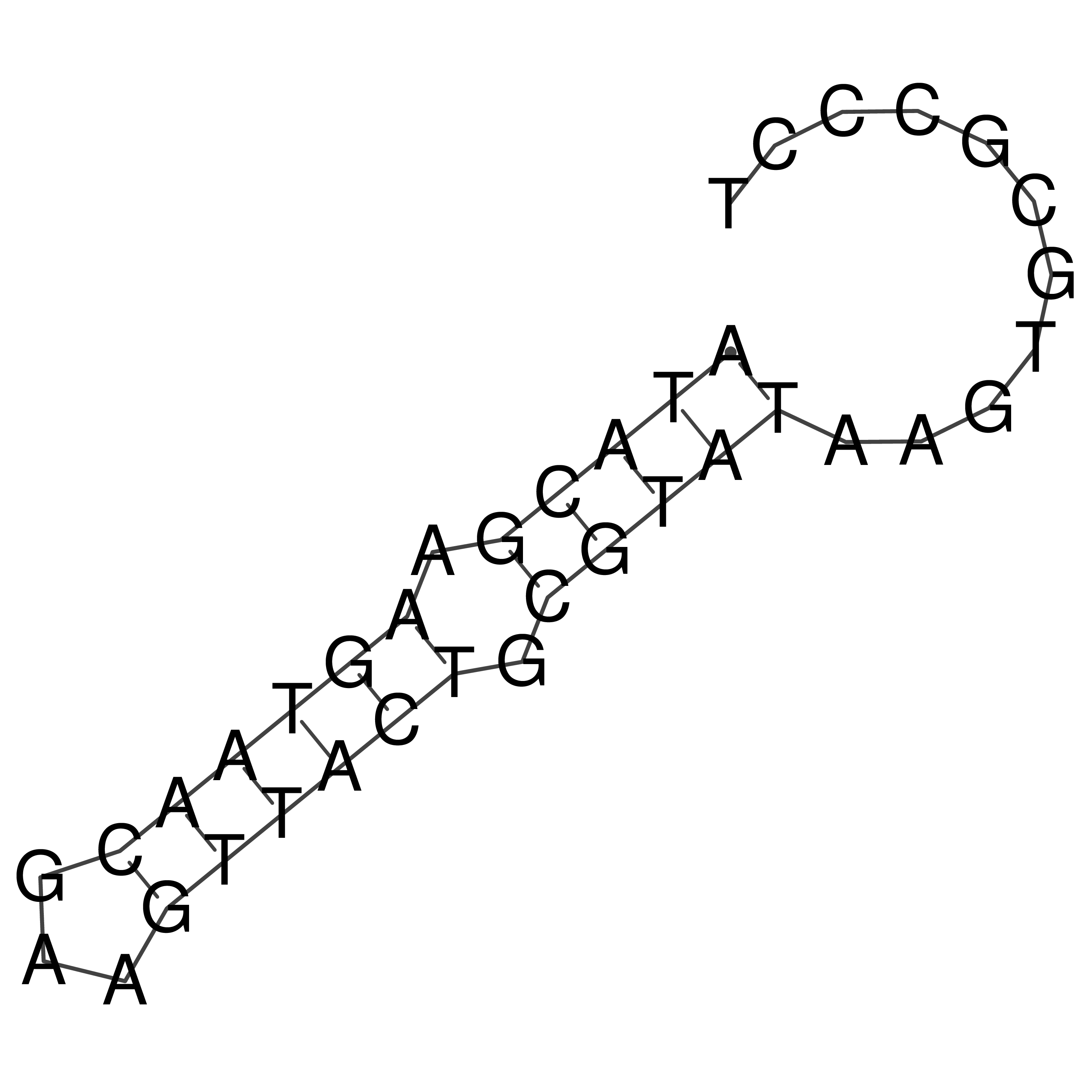
Reference
[1] Kurenbach B et al. (2006) The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology. 152(Pt 3):637-45. [PMID:16514144]
[2] Kurenbach B et al. (2003) Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 50(1):86-93. [PMID:12826062]
[3] Wang A et al. (1995) Streptococcal Plasmid pIP501 Has a Functional oriT Site. J Bacteriol. 177(15):4199-206. [PMID:7635806]
Relaxase
| ID | 5 | GenBank | AAA99466 |
| Name | TraA_pIP501 |
UniProt ID | Q52001 |
| Length | 654 a.a. | PDB ID | |
| Note | (1) The N-terminal domin (246 amino acids) of relaxase TraA_pIP501 show specific oriT binding [PMID:15554903]. (2) The relaxase TraA_pIP501 and its N-terminal domin (246 amino acids) behave as dimers in solution [PMID:15554903]. (3) The relaxase TraA_pIP501 negatively autoregulate the downstream 15 tra gene cluster [PMID:16514144]. |
||
Relaxase protein sequence
Download Length: 654 a.a. Molecular weight: 76493.35 Da Isoelectric Point: 9.3594
MTIAKRENGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTLWNKMELA
EKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVNEGMIADVAIHRDDENNPHAHIMLTMREVDSE
GNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNDWGRKSLVSEIRKDWADKVNQYLKDRNIDQQITEK
SHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLENQEKVLKQDQNFTLKFEK
TFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERDMLTKANE
ILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKKIFKEKPF
QTSRYLDSKIKQIEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDSVIEGEML
LAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHIDELSPLAK
QNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSGNQLQKKR
NLDKLIKQTKAKKNQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q52001 |
Reference
[1] Kurenbach B et al. (2006) The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology. 152(Pt 3):637-45. [PMID:16514144]
[2] Kurenbach B et al. (2003) Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 50(1):86-93. [PMID:12826062]
Host bacterium
| ID | 5 | GenBank | L39769 |
| Plasmid name | pIP501 | Incompatibility group | Inc18 |
| Plasmid size | 8136 bp | Coordinate of oriT [Strand] | 1259..1296 [-] |
| Host baterium | Streptococcus agalactiae B-49813/75 |
Cargo genes
| Drug resistance gene | resitance to chloramphenicol and MLS (macrolide, lincosamide, streptogramin B antibiotics) |
| Virulence gene | _ |
| Metal resistance gene | _ |
| Degradation gene | _ |
| Symbiosis gene | - |
| Anti-CRISPR | - |