
| Conjugative transfers of single-stranded DNA (ssDNA) |
|
Conjugative transfers of plasmids and chromosome-borne mobile genetic elements can be devided into two different conjugative transfer concepts,
single-stranded DNA (ssDNA) transfer and double-stranded (dsDNA) transfer [Goessweiner-Mohr, et al (2013) Plasmid]. The most widespread strategy used by plasmids/mobile elements of both
Gram-negative and Gram-positive bacteria relies on the translocation of an ssDNA molecule through a type IV secretion system (T4SS) [Carraro, et al (2014) Microbiol Spectr;
Bellanger, et al (2014) FEMS Microbiol Rev].
The prominent feature of ssDNA transfer is that a DNA single strand is generated by the recognition and cleavage of the relaxase at the nic site of oriT [Grohmann, et al (2003) Microbiol Mol Biol Rev].
|
| oriT, relaxosome, T4CP |
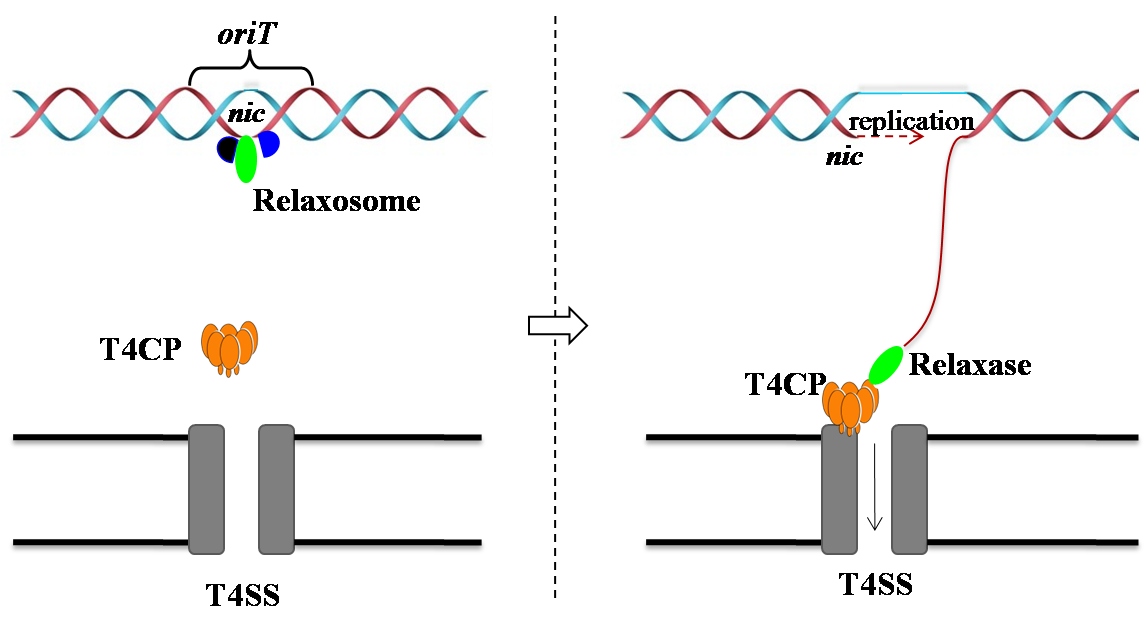 |
|
(1) Origin of transfer (oriT) is a stretch of DNA usually of several hundred base pairs (bp) in length.
Within oriT is the nic site where the relaxase, a phosphodiesterase, cleaves in a site- and strand-specific manner,
resulting in the transfer of a ssDNA in a 5'-3' direction to the recipient cell. The nic site undergoes
cleavage and religation during vegetative growth, but this is converted to a cleavage and unwinding reaction
when a competent mating pair has formed [de la Cruz, et al (2010) FEMS Microbiol Rev].
(2) The relaxosome, a multiprotein-DNA complex formed at the origin of transfer (oriT), consists of a relaxase, directed to the nic site by auxiliary DNA-binding proteins [de la Cruz, et al (2010) FEMS Microbiol Rev; Grohmann, et al (2003) Microbiol Mol Biol Rev]. (3) The coupling protein (T4CP) guides this protein-DNA (relaxosome-oriT) complex to the T4SS at the membrane, where the transferred strand gets out of the bacteria [Llosa, et al (2002) Mol Microbiol]. |
| oriTDB |
 Length distributions of 238 oriTs from natural plasmids and ICEs/IMEs collected by text mining (February 15th, 2017).
The web-based oriTDB database was designed to facilitate the identification and functional interpretation
of conjugative transfers of ssDNA of Gram-negative or Gram-positive bacteria.
It currently contains details of transfer origin regions (oriT), nicking sites (nic), relaxases
and auxiliary DNA-binding proteins, type IV coupling proteins (T4CP),
as well as a collection of directly related references.
The congruent relationship between oriT and relaxosome, correlation between relaxase and auxiliary DNA-binding protein(s), and connection between oriT-relaxosome complex and the T4CP were displayed in the oriTDB. Users can create genetic maps of oriTs and investigate the inverted repeat (IR) sequences and the nic site recognized by the relaxase with the embedded graphic display. |