Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 37371..37635 | Replicon | plasmid p266917_2_03 |
| Accession | NZ_CP026726 | ||
| Organism | Escherichia coli strain 266917_2 | ||
Toxin (Protein)
| Gene name | pndA | Uniprot ID | U9Z417 |
| Locus tag | C4R22_RS30465 | Protein ID | WP_001387489.1 |
| Coordinates | 37483..37635 (+) | Length | 51 a.a. |
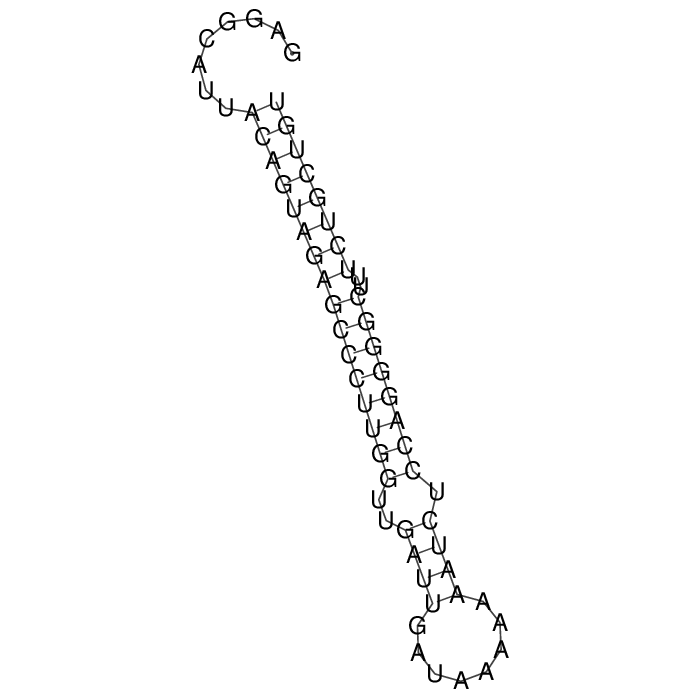
Antitoxin (RNA)
| Gene name | pndB | ||
| Locus tag | - | ||
| Coordinates | 37371..37431 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| C4R22_RS30450 | 33473..34543 | - | 1071 | WP_011264050.1 | IncI1-type conjugal transfer protein TrbB | - |
| C4R22_RS30455 | 34562..35770 | - | 1209 | WP_011264049.1 | IncI1-type conjugal transfer protein TrbA | - |
| - | 35950..36007 | - | 58 | NuclAT_1 | - | - |
| - | 35950..36007 | - | 58 | NuclAT_1 | - | - |
| - | 35950..36007 | - | 58 | NuclAT_1 | - | - |
| - | 35950..36007 | - | 58 | NuclAT_1 | - | - |
| C4R22_RS30460 | 36077..37168 | - | 1092 | WP_069067420.1 | protein finQ | - |
| - | 37371..37431 | - | 61 | NuclAT_0 | - | Antitoxin |
| - | 37371..37431 | - | 61 | NuclAT_0 | - | Antitoxin |
| - | 37371..37431 | - | 61 | NuclAT_0 | - | Antitoxin |
| - | 37371..37431 | - | 61 | NuclAT_0 | - | Antitoxin |
| C4R22_RS30465 | 37483..37635 | + | 153 | WP_001387489.1 | Hok/Gef family protein | Toxin |
| C4R22_RS30470 | 37707..37958 | - | 252 | WP_001291964.1 | hypothetical protein | - |
| C4R22_RS30475 | 38258..38554 | + | 297 | WP_011264046.1 | hypothetical protein | - |
| C4R22_RS31970 | 38619..38795 | - | 177 | WP_001054897.1 | hypothetical protein | - |
| C4R22_RS30480 | 39187..39396 | + | 210 | WP_000062603.1 | HEAT repeat domain-containing protein | - |
| C4R22_RS30485 | 39468..40130 | - | 663 | WP_000644796.1 | plasmid IncI1-type surface exclusion protein ExcA | - |
| C4R22_RS30490 | 40201..42369 | - | 2169 | WP_023518847.1 | DotA/TraY family protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | - | - | 1..83010 | 83010 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 51 a.a. Molecular weight: 5761.10 Da Isoelectric Point: 8.7948
>T95599 WP_001387489.1 NZ_CP026726:37483-37635 [Escherichia coli]
MPQRTFLMMLIVVCVTILCFVWMVRDSLCGLRLQQGNTVLVATLAYEVKR
MPQRTFLMMLIVVCVTILCFVWMVRDSLCGLRLQQGNTVLVATLAYEVKR
Download Length: 153 bp
>T95599 NZ_CP026726:37483-37635 [Escherichia coli]
ATGCCACAGCGAACGTTTTTAATGATGTTAATCGTCGTCTGTGTGACGATTCTGTGTTTTGTCTGGATGGTGAGGGATTC
GCTTTGCGGACTCCGGCTCCAGCAGGGAAACACAGTGCTTGTGGCAACGTTAGCCTACGAAGTTAAACGTTAA
ATGCCACAGCGAACGTTTTTAATGATGTTAATCGTCGTCTGTGTGACGATTCTGTGTTTTGTCTGGATGGTGAGGGATTC
GCTTTGCGGACTCCGGCTCCAGCAGGGAAACACAGTGCTTGTGGCAACGTTAGCCTACGAAGTTAAACGTTAA
Antitoxin
Download Length: 61 bp
>AT95599 NZ_CP026726:c37431-37371 [Escherichia coli]
GAGGCATTACAGTAGAGCCCTTGGTTGATTGATAAAAAAATCTCCAGGGGCTTTCTGCTGT
GAGGCATTACAGTAGAGCCCTTGGTTGATTGATAAAAAAATCTCCAGGGGCTTTCTGCTGT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|