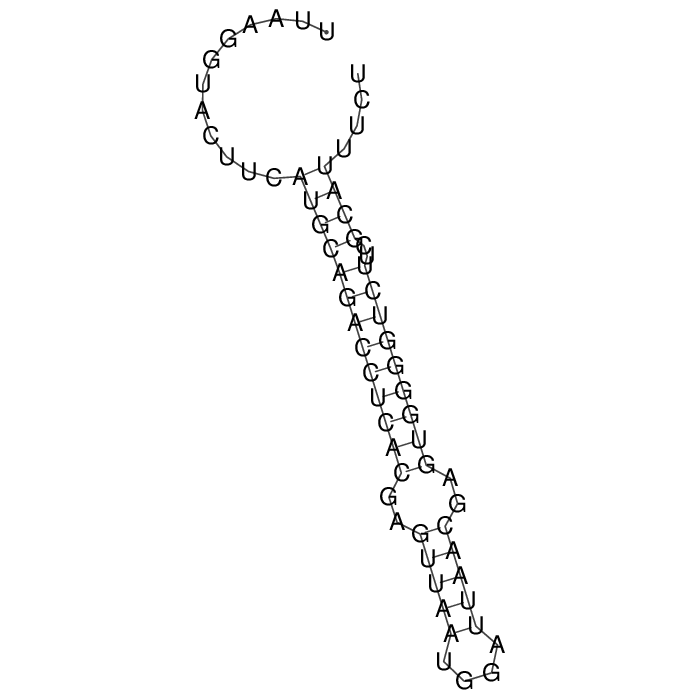
Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 73613..73867 | Replicon | plasmid pCREC-544_1 |
| Accession | NZ_CP024827 | ||
| Organism | Escherichia coli strain CREC-544 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | - |
| Locus tag | CU077_RS25205 | Protein ID | WP_001351576.1 |
| Coordinates | 73613..73819 (-) | Length | 69 a.a. |
Antitoxin (RNA)
| Gene name | sok | ||
| Locus tag | - | ||
| Coordinates | 73806..73867 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CU077_RS25165 | 69165..69566 | - | 402 | WP_001398199.1 | type II toxin-antitoxin system toxin endoribonuclease PemK | - |
| CU077_RS26320 | 69499..69756 | - | 258 | WP_000557619.1 | type II toxin-antitoxin system antitoxin PemI | - |
| CU077_RS25170 | 69849..70502 | - | 654 | WP_000616807.1 | CPBP family intramembrane metalloprotease | - |
| CU077_RS26460 | 70600..70740 | - | 141 | WP_001333237.1 | hypothetical protein | - |
| CU077_RS25180 | 71441..72298 | - | 858 | WP_000774297.1 | incFII family plasmid replication initiator RepA | - |
| CU077_RS25185 | 72291..72773 | - | 483 | WP_001273588.1 | hypothetical protein | - |
| CU077_RS25190 | 72766..72840 | - | 75 | WP_001442103.1 | RepA leader peptide Tap | - |
| CU077_RS25200 | 73072..73329 | - | 258 | WP_000083845.1 | replication regulatory protein RepA | - |
| CU077_RS25205 | 73613..73819 | - | 207 | WP_001351576.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 73806..73867 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 73806..73867 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 73806..73867 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 73806..73867 | + | 62 | NuclAT_1 | - | Antitoxin |
| CU077_RS26465 | 74123..74197 | - | 75 | Protein_87 | endonuclease | - |
| CU077_RS25215 | 74443..74655 | - | 213 | WP_001299730.1 | ANR family transcriptional regulator | - |
| CU077_RS25220 | 74791..75351 | - | 561 | WP_000139315.1 | fertility inhibition protein FinO | - |
| CU077_RS25225 | 75454..76314 | - | 861 | WP_000704513.1 | alpha/beta hydrolase | - |
| CU077_RS25230 | 76373..77119 | - | 747 | WP_000205749.1 | conjugal transfer pilus acetylase TraX | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | sitABCD / aph(3')-Ia / catA2 / dfrA14 / sul2 / aph(3'')-Ib / aph(6)-Id / blaTEM-1B / blaCTX-M-55 | iucC / iucB / iucA / iutA / iucD | 1..122937 | 122937 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 69 a.a. Molecular weight: 7826.31 Da Isoelectric Point: 8.8807
>T88700 WP_001351576.1 NZ_CP024827:c73819-73613 [Escherichia coli]
MKYLNTTDCSLFFAERSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MKYLNTTDCSLFFAERSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 207 bp
>T88700 NZ_CP024827:c73819-73613 [Escherichia coli]
ATGAAGTACCTTAACACTACTGATTGTAGCCTCTTCTTTGCTGAGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTATGTGAGCTGAATATTCACAGAG
GAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
ATGAAGTACCTTAACACTACTGATTGTAGCCTCTTCTTTGCTGAGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTATGTGAGCTGAATATTCACAGAG
GAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
Antitoxin
Download Length: 62 bp
>AT88700 NZ_CP024827:73806-73867 [Escherichia coli]
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|