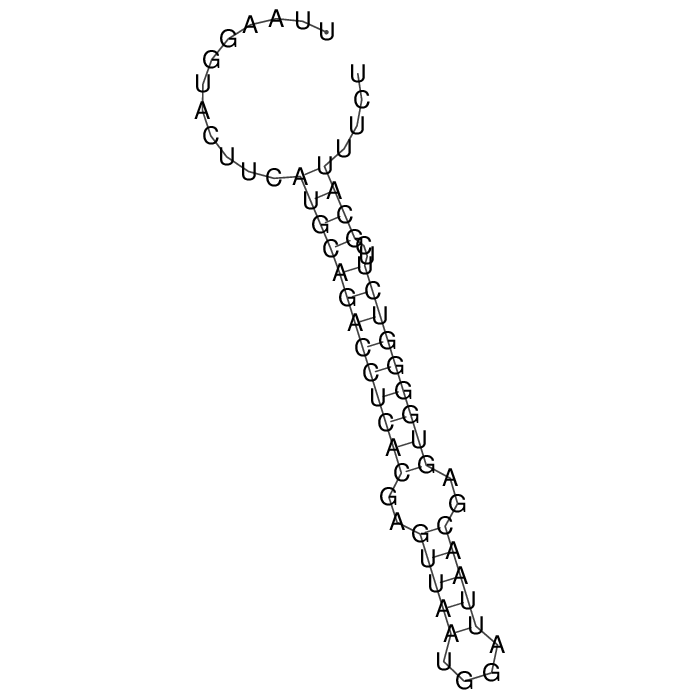
Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 125668..125922 | Replicon | plasmid pCREC-629_1 |
| Accession | NZ_CP024816 | ||
| Organism | Escherichia coli strain CREC-629 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | - |
| Locus tag | CU080_RS26530 | Protein ID | WP_015387399.1 |
| Coordinates | 125668..125874 (-) | Length | 69 a.a. |
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 125861..125922 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CU080_RS26495 | 120915..121826 | - | 912 | WP_000440183.1 | carbamate kinase | - |
| CU080_RS26500 | 121837..123057 | - | 1221 | WP_100183869.1 | arginine deiminase | - |
| CU080_RS26505 | 123950..124432 | + | 483 | Protein_145 | VENN motif pre-toxin domain-containing protein | - |
| CU080_RS26510 | 124378..124824 | - | 447 | Protein_146 | incFII family plasmid replication initiator RepA | - |
| CU080_RS26515 | 124817..124891 | - | 75 | WP_001365705.1 | RepA leader peptide Tap | - |
| CU080_RS26525 | 125127..125384 | - | 258 | WP_000083833.1 | replication regulatory protein RepA | - |
| CU080_RS26530 | 125668..125874 | - | 207 | WP_015387399.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 125861..125922 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 125861..125922 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 125861..125922 | + | 62 | NuclAT_1 | - | Antitoxin |
| - | 125861..125922 | + | 62 | NuclAT_1 | - | Antitoxin |
| CU080_RS26540 | 126061..126243 | - | 183 | WP_000968309.1 | hypothetical protein | - |
| CU080_RS26550 | 126344..127046 | + | 703 | Protein_151 | IS1 family transposase | - |
| CU080_RS26560 | 127347..127559 | - | 213 | WP_005012601.1 | hypothetical protein | - |
| CU080_RS26565 | 127692..128252 | - | 561 | WP_000139363.1 | fertility inhibition protein FinO | - |
| CU080_RS26570 | 128307..129053 | - | 747 | WP_000205701.1 | conjugal transfer pilus acetylase TraX | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | aac(3)-IId / tet(B) / erm(B) / mph(A) / sul1 / qacE / aadA5 / dfrA17 / blaTEM-1B | - | 1..176274 | 176274 | |
| - | flank | IS/Tn | - | - | 126660..127046 | 386 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 69 a.a. Molecular weight: 7720.23 Da Isoelectric Point: 9.3127
>T88611 WP_015387399.1 NZ_CP024816:c125874-125668 [Escherichia coli]
MKYLNTTDCSLFLAGRSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MKYLNTTDCSLFLAGRSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 207 bp
>T88611 NZ_CP024816:c125874-125668 [Escherichia coli]
ATGAAGTACCTTAACACTACTGATTGTAGCCTCTTCCTTGCAGGGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTATGTGAACTGAATATTCACAGGG
GAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
ATGAAGTACCTTAACACTACTGATTGTAGCCTCTTCCTTGCAGGGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTATGTGAACTGAATATTCACAGGG
GAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
Antitoxin
Download Length: 62 bp
>AT88611 NZ_CP024816:125861-125922 [Escherichia coli]
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|