Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | ldrD-rdlD/Ldr(toxin) |
| Location | 1377246..1377468 | Replicon | chromosome |
| Accession | NZ_CP024275 | ||
| Organism | Escherichia coli O6:H16 strain M9682-C1 | ||
Toxin (Protein)
| Gene name | ldrD | Uniprot ID | B7LGX8 |
| Locus tag | COG40_RS07185 | Protein ID | WP_000170965.1 |
| Coordinates | 1377246..1377353 (-) | Length | 36 a.a. |
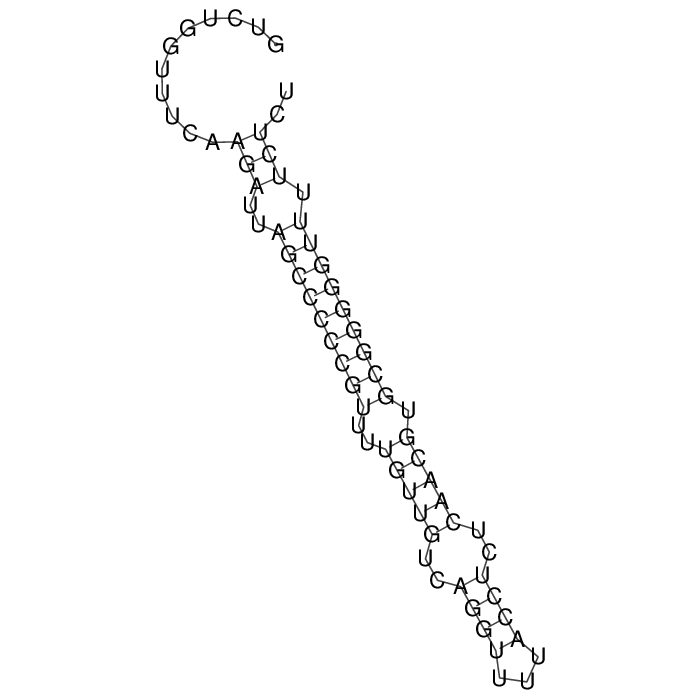
Antitoxin (RNA)
| Gene name | rdlD | ||
| Locus tag | - | ||
| Coordinates | 1377401..1377468 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| COG40_RS07160 | 1373092..1374174 | + | 1083 | WP_000804732.1 | peptide chain release factor 1 | - |
| COG40_RS07165 | 1374174..1375007 | + | 834 | WP_000456454.1 | peptide chain release factor N(5)-glutamine methyltransferase | - |
| COG40_RS07170 | 1375004..1375396 | + | 393 | WP_001705045.1 | invasion regulator SirB2 | - |
| COG40_RS07175 | 1375400..1376209 | + | 810 | WP_001257044.1 | invasion regulator SirB1 | - |
| COG40_RS07180 | 1376245..1377099 | + | 855 | WP_000811065.1 | 3-deoxy-8-phosphooctulonate synthase | - |
| COG40_RS07185 | 1377246..1377353 | - | 108 | WP_000170965.1 | type I toxin-antitoxin system toxin Ldr family protein | Toxin |
| - | 1377401..1377468 | + | 68 | NuclAT_14 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_14 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_14 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_14 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_16 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_16 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_16 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_16 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_18 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_18 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_18 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_18 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_20 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_20 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_20 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_20 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_22 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_22 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_22 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_22 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_24 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_24 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_24 | - | Antitoxin |
| - | 1377401..1377468 | + | 68 | NuclAT_24 | - | Antitoxin |
| COG40_RS07190 | 1377781..1377888 | - | 108 | WP_000170963.1 | small toxic polypeptide LdrB | - |
| - | 1377936..1378002 | + | 67 | NuclAT_26 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_26 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_26 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_26 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_27 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_27 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_27 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_27 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_28 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_28 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_28 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_28 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_29 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_29 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_29 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_29 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_30 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_30 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_30 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_30 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_31 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_31 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_31 | - | - |
| - | 1377936..1378002 | + | 67 | NuclAT_31 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_15 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_15 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_15 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_15 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_17 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_17 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_17 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_17 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_19 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_19 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_19 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_19 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_21 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_21 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_21 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_21 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_23 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_23 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_23 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_23 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_25 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_25 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_25 | - | - |
| - | 1377938..1378003 | + | 66 | NuclAT_25 | - | - |
| COG40_RS07195 | 1378293..1379393 | - | 1101 | WP_001295620.1 | sodium-potassium/proton antiporter ChaA | - |
| COG40_RS07200 | 1379663..1379893 | + | 231 | WP_001146444.1 | putative cation transport regulator ChaB | - |
| COG40_RS07205 | 1380051..1380746 | + | 696 | WP_001297117.1 | glutathione-specific gamma-glutamylcyclotransferase | - |
| COG40_RS07210 | 1380790..1381143 | - | 354 | WP_001169669.1 | DsrE/F sulfur relay family protein YchN | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 36 a.a. Molecular weight: 4001.77 Da Isoelectric Point: 11.4779
>T87465 WP_000170965.1 NZ_CP024275:c1377353-1377246 [Escherichia coli O6:H16]
MTLAQFAMTFWHDLAAPILAGIITAAIVSWWRNRK
MTLAQFAMTFWHDLAAPILAGIITAAIVSWWRNRK
Download Length: 108 bp
>T87465 NZ_CP024275:c1377353-1377246 [Escherichia coli O6:H16]
ATGACGCTCGCGCAGTTTGCCATGACTTTCTGGCACGACCTGGCAGCACCGATCCTGGCGGGGATTATTACCGCAGCGAT
TGTCAGCTGGTGGCGTAACCGGAAGTAA
ATGACGCTCGCGCAGTTTGCCATGACTTTCTGGCACGACCTGGCAGCACCGATCCTGGCGGGGATTATTACCGCAGCGAT
TGTCAGCTGGTGGCGTAACCGGAAGTAA
Antitoxin
Download Length: 68 bp
>AT87465 NZ_CP024275:1377401-1377468 [Escherichia coli O6:H16]
GTCTGGTTTCAAGATTAGCCCCCGTTTTGTTGTCAGGTTTTACCTCTCAACGTGCGGGGGTTTTCTCT
GTCTGGTTTCAAGATTAGCCCCCGTTTTGTTGTCAGGTTTTACCTCTCAACGTGCGGGGGTTTTCTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|