Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 70265..70504 | Replicon | plasmid p2_0.1 |
| Accession | NZ_CP023854 | ||
| Organism | Escherichia coli strain 2/0 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | A0A762TWR7 |
| Locus tag | CRT29_RS26260 | Protein ID | WP_023144756.1 |
| Coordinates | 70370..70504 (+) | Length | 45 a.a. |
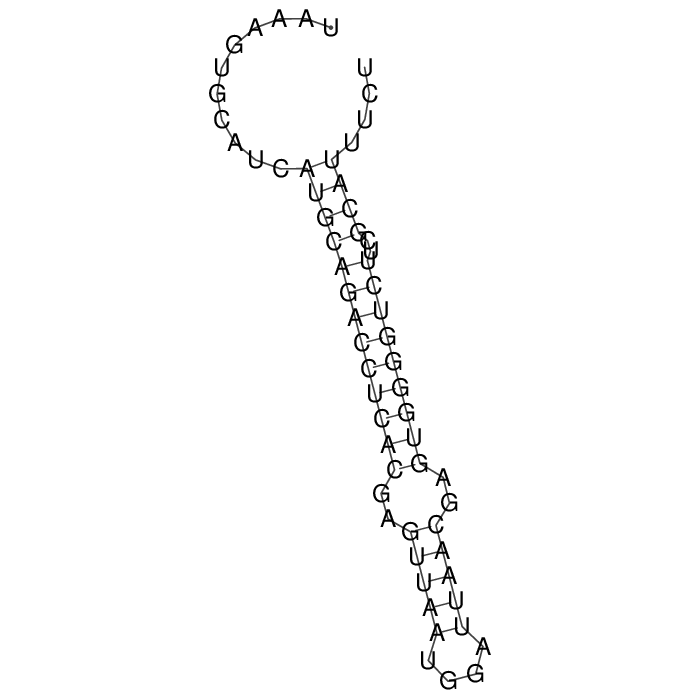
Antitoxin (RNA)
| Gene name | sok | ||
| Locus tag | - | ||
| Coordinates | 70265..70325 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CRT29_RS26235 | 67795..68541 | + | 747 | WP_000205718.1 | conjugal transfer pilus acetylase TraX | - |
| CRT29_RS26240 | 68596..69156 | + | 561 | WP_000139341.1 | fertility inhibition protein FinO | - |
| CRT29_RS26245 | 69287..69499 | + | 213 | WP_013023861.1 | hypothetical protein | - |
| CRT29_RS26860 | 70012..70298 | + | 287 | Protein_84 | DUF2726 domain-containing protein | - |
| - | 70265..70325 | - | 61 | NuclAT_2 | - | Antitoxin |
| - | 70265..70325 | - | 61 | NuclAT_2 | - | Antitoxin |
| - | 70265..70325 | - | 61 | NuclAT_2 | - | Antitoxin |
| - | 70265..70325 | - | 61 | NuclAT_2 | - | Antitoxin |
| CRT29_RS26260 | 70370..70504 | + | 135 | WP_023144756.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| CRT29_RS26265 | 70801..71055 | + | 255 | WP_000083850.1 | replication regulatory protein RepA | - |
| CRT29_RS26270 | 71215..71961 | - | 747 | WP_001016257.1 | IS21-like element ISEc12 family helper ATPase IstB | - |
| CRT29_RS26275 | 71976..73517 | - | 1542 | WP_002431311.1 | IS21-like element ISEc12 family transposase | - |
| CRT29_RS26980 | 73748..73882 | + | 135 | Protein_89 | protein CopA/IncA | - |
| CRT29_RS26285 | 73879..73953 | + | 75 | WP_014966221.1 | RepA leader peptide Tap | - |
| CRT29_RS26290 | 73946..74803 | + | 858 | WP_000130646.1 | incFII family plasmid replication initiator RepA | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | aadA5 / qacE / sul1 / mph(A) / blaTEM-1B / tet(B) | - | 1..135053 | 135053 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 45 a.a. Molecular weight: 4896.90 Da Isoelectric Point: 7.7209
>T86305 WP_023144756.1 NZ_CP023854:70370-70504 [Escherichia coli]
MTKYTLIGLLAVCATVLCFSLIFREQLCELNIHRGNTVVQVTLA
MTKYTLIGLLAVCATVLCFSLIFREQLCELNIHRGNTVVQVTLA
Download Length: 135 bp
>T86305 NZ_CP023854:70370-70504 [Escherichia coli]
ATGACGAAATATACCCTTATCGGGTTGCTTGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACAGTT
ATGTGAACTGAATATTCACAGGGGAAATACAGTGGTGCAGGTAACTCTGGCCTGA
ATGACGAAATATACCCTTATCGGGTTGCTTGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACAGTT
ATGTGAACTGAATATTCACAGGGGAAATACAGTGGTGCAGGTAACTCTGGCCTGA
Antitoxin
Download Length: 61 bp
>AT86305 NZ_CP023854:c70325-70265 [Escherichia coli]
TAAAGTGCATCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TAAAGTGCATCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|