Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 137908..138161 | Replicon | plasmid pCFSAN004181P |
| Accession | NZ_CP012499 | ||
| Organism | Escherichia coli strain GB089 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | G9G1E3 |
| Locus tag | CO57_RS00710 | Protein ID | WP_001312851.1 |
| Coordinates | 137908..138057 (-) | Length | 50 a.a. |
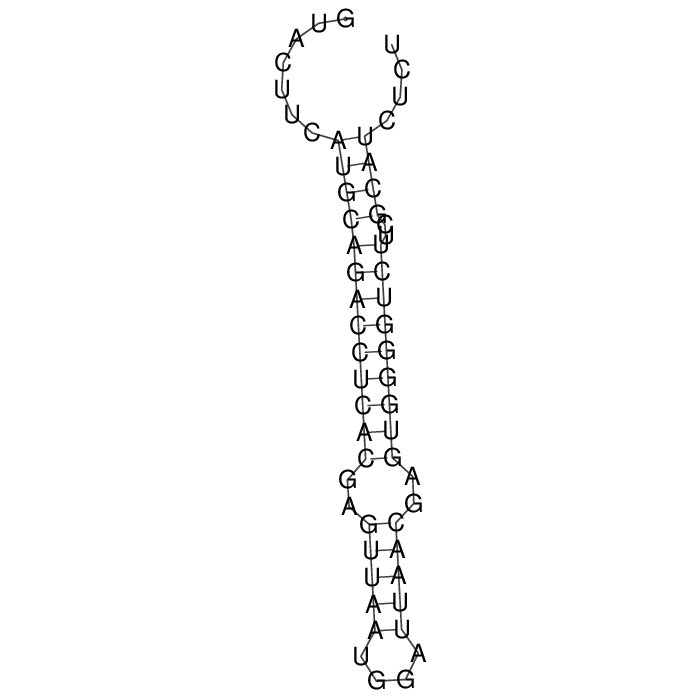
Antitoxin (RNA)
| Gene name | sok | ||
| Locus tag | - | ||
| Coordinates | 138105..138161 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CO57_RS00680 | 133424..133888 | - | 465 | WP_040090730.1 | hypothetical protein | - |
| CO57_RS00685 | 134167..135207 | - | 1041 | WP_136759863.1 | hypothetical protein | - |
| CO57_RS00695 | 136208..137065 | - | 858 | WP_000829162.1 | incFII family plasmid replication initiator RepA | - |
| CO57_RS00700 | 137058..137132 | - | 75 | WP_001364037.1 | RepA leader peptide Tap | - |
| CO57_RS00705 | 137376..137624 | - | 249 | WP_000083838.1 | replication regulatory protein RepA | - |
| CO57_RS00710 | 137908..138057 | - | 150 | WP_001312851.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 138105..138161 | + | 57 | NuclAT_1 | - | Antitoxin |
| - | 138105..138161 | + | 57 | NuclAT_1 | - | Antitoxin |
| - | 138105..138161 | + | 57 | NuclAT_1 | - | Antitoxin |
| - | 138105..138161 | + | 57 | NuclAT_1 | - | Antitoxin |
| CO57_RS00715 | 138313..138777 | - | 465 | WP_000455478.1 | hypothetical protein | - |
| CO57_RS00720 | 138932..139522 | - | 591 | WP_000766788.1 | DUF2726 domain-containing protein | - |
| CO57_RS00725 | 139560..139769 | - | 210 | WP_001355319.1 | hemolysin expression modulator Hha | - |
| CO57_RS00730 | 140097..140309 | - | 213 | WP_001364091.1 | hypothetical protein | - |
| CO57_RS00735 | 140444..141004 | - | 561 | WP_000139355.1 | fertility inhibition protein FinO | - |
| CO57_RS00740 | 141059..141796 | - | 738 | WP_000429592.1 | conjugal transfer pilus acetylase TraX | - |
| CO57_RS00745 | 141818..142249 | - | 432 | WP_000447372.1 | hypothetical protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | - | faeJ / faeI / faeH / faeF / faeE / faeD / faeC / hlyD / hlyB / hlyA / hlyC | 1..223952 | 223952 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 50 a.a. Molecular weight: 5542.67 Da Isoelectric Point: 8.7678
>T56501 WP_001312851.1 NZ_CP012499:c138057-137908 [Escherichia coli]
MTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 150 bp
>T56501 NZ_CP012499:c138057-137908 [Escherichia coli]
ATGACGAAATATGCCCTTATCGGGTTGCTCGCTGTGTGCGCTACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTT
ATGTGAACTGAATATTCACAGAGGAAATACAGTGGTGCAGGTAACTTTGGCCTACGAAGCACGGAAGTAA
ATGACGAAATATGCCCTTATCGGGTTGCTCGCTGTGTGCGCTACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTT
ATGTGAACTGAATATTCACAGAGGAAATACAGTGGTGCAGGTAACTTTGGCCTACGAAGCACGGAAGTAA
Antitoxin
Download Length: 57 bp
>AT56501 NZ_CP012499:138105-138161 [Escherichia coli]
GTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATCTCT
GTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATCTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|