Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | symER/SymE(toxin) |
| Location | 5292511..5292923 | Replicon | chromosome |
| Accession | NZ_AP025747 | ||
| Organism | Escherichia sp. HH091_1A strain HH-P024 | ||
Toxin (Protein)
| Gene name | symE | Uniprot ID | A0A8D9PK47 |
| Locus tag | QUF14_RS26150 | Protein ID | WP_000132619.1 |
| Coordinates | 5292511..5292852 (-) | Length | 114 a.a. |
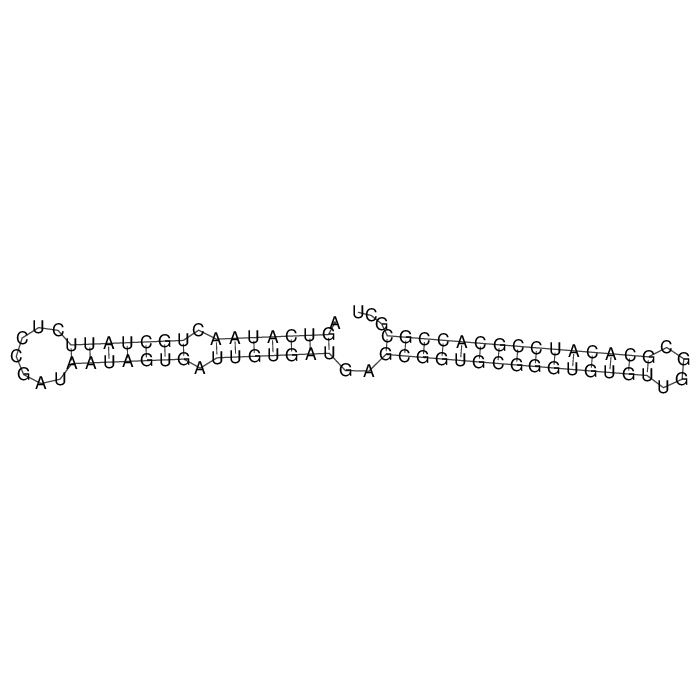
Antitoxin (RNA)
| Gene name | symR | ||
| Locus tag | - | ||
| Coordinates | 5292847..5292923 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QUF14_RS26130 (5288521) | 5288521..5289933 | - | 1413 | WP_000199312.1 | PLP-dependent aminotransferase family protein | - |
| QUF14_RS26135 (5290110) | 5290110..5290274 | + | 165 | WP_000394281.1 | DUF1127 domain-containing protein | - |
| QUF14_RS26140 (5290371) | 5290371..5291253 | + | 883 | Protein_5097 | DUF262 domain-containing protein | - |
| QUF14_RS26145 (5291301) | 5291301..5292464 | + | 1164 | WP_138983873.1 | DUF1524 domain-containing protein | - |
| QUF14_RS26150 (5292511) | 5292511..5292852 | - | 342 | WP_000132619.1 | endoribonuclease SymE | Toxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_23 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_23 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_23 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_23 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_24 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_24 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_24 | - | Antitoxin |
| - (5292847) | 5292847..5292923 | + | 77 | NuclAT_24 | - | Antitoxin |
| QUF14_RS26155 (5293073) | 5293073..5294824 | - | 1752 | WP_000110091.1 | restriction endonuclease subunit S | - |
| QUF14_RS26160 (5294824) | 5294824..5296293 | - | 1470 | WP_001387312.1 | type I restriction-modification system subunit M | - |
| QUF14_RS26165 (5296360) | 5296360..5297229 | - | 870 | Protein_5102 | type I restriction-modification enzyme R subunit C-terminal domain-containing protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 114 a.a. Molecular weight: 12322.13 Da Isoelectric Point: 7.8219
>T40767 WP_000132619.1 NZ_AP025747:c5292852-5292511 [Escherichia sp. HH091_1A]
MTDTHSIAQPFEAEVSPANNRQLTVSYASRYPDYSRIPAITLKGQWLEAAGFATGTAVDVKVMEGCIVLTAQPPAPEESE
LMQSLRQVCKLSARKQKQVQEFIGVIAGKQKVA
MTDTHSIAQPFEAEVSPANNRQLTVSYASRYPDYSRIPAITLKGQWLEAAGFATGTAVDVKVMEGCIVLTAQPPAPEESE
LMQSLRQVCKLSARKQKQVQEFIGVIAGKQKVA
Download Length: 342 bp
>T40767 NZ_AP025747:c5292852-5292511 [Escherichia sp. HH091_1A]
ATGACTGACACGCATTCTATTGCACAACCGTTCGAAGCAGAAGTCTCCCCGGCAAATAACCGTCAATTAACCGTGAGTTA
TGCGAGTCGCTACCCGGATTACAGCCGTATTCCCGCCATCACCCTGAAAGGTCAGTGGCTGGAAGCCGCCGGTTTTGCCA
CCGGCACGGCGGTAGATGTCAAAGTGATGGAAGGCTGTATTGTCCTCACCGCCCAACCACCCGCCCCCGAAGAGAGCGAA
CTGATGCAGTCGCTGCGCCAGGTGTGCAAACTGTCAGCGCGTAAACAGAAGCAGGTGCAGGAGTTTATTGGGGTGATTGC
CGGTAAACAGAAAGTCGCCTGA
ATGACTGACACGCATTCTATTGCACAACCGTTCGAAGCAGAAGTCTCCCCGGCAAATAACCGTCAATTAACCGTGAGTTA
TGCGAGTCGCTACCCGGATTACAGCCGTATTCCCGCCATCACCCTGAAAGGTCAGTGGCTGGAAGCCGCCGGTTTTGCCA
CCGGCACGGCGGTAGATGTCAAAGTGATGGAAGGCTGTATTGTCCTCACCGCCCAACCACCCGCCCCCGAAGAGAGCGAA
CTGATGCAGTCGCTGCGCCAGGTGTGCAAACTGTCAGCGCGTAAACAGAAGCAGGTGCAGGAGTTTATTGGGGTGATTGC
CGGTAAACAGAAAGTCGCCTGA
Antitoxin
Download Length: 77 bp
>AT40767 NZ_AP025747:5292847-5292923 [Escherichia sp. HH091_1A]
AGTCATAACTGCTATTCTCCGATAATAGTGATTGTGATGAGCGGTGCGGGTGTGTTGGCGCACATCCGCACCGCGCT
AGTCATAACTGCTATTCTCCGATAATAGTGATTGTGATGAGCGGTGCGGGTGTGTTGGCGCACATCCGCACCGCGCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|