Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 85032..85286 | Replicon | plasmid pMTY13867_IncFI |
| Accession | NZ_AP025658 | ||
| Organism | Escherichia coli strain TUM13867 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | G9G1E3 |
| Locus tag | MWM17_RS23875 | Protein ID | WP_001312851.1 |
| Coordinates | 85032..85181 (-) | Length | 50 a.a. |
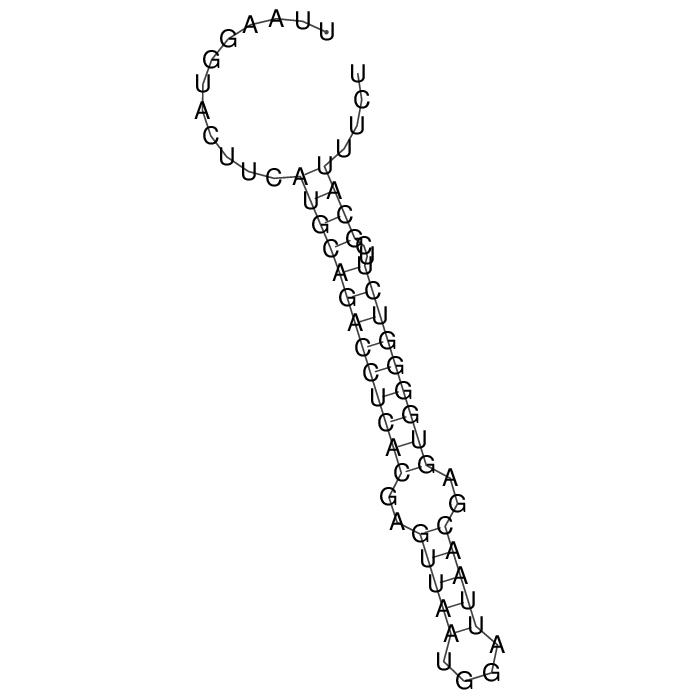
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 85225..85286 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MWM17_RS23845 (80314) | 80314..83319 | + | 3006 | WP_001143760.1 | Tn3-like element Tn3 family transposase | - |
| MWM17_RS23850 (83252) | 83252..83717 | - | 466 | Protein_75 | plasmid replication initiator RepA | - |
| MWM17_RS23855 (83710) | 83710..84192 | - | 483 | WP_001273588.1 | hypothetical protein | - |
| MWM17_RS23860 (84185) | 84185..84232 | - | 48 | WP_229471593.1 | hypothetical protein | - |
| MWM17_RS23865 (84223) | 84223..84474 | + | 252 | WP_223195197.1 | replication protein RepA | - |
| MWM17_RS23870 (84491) | 84491..84748 | - | 258 | WP_000083845.1 | replication regulatory protein RepA | - |
| MWM17_RS23875 (85032) | 85032..85181 | - | 150 | WP_001312851.1 | Hok/Gef family protein | Toxin |
| - (85225) | 85225..85286 | + | 62 | NuclAT_1 | - | Antitoxin |
| - (85225) | 85225..85286 | + | 62 | NuclAT_1 | - | Antitoxin |
| - (85225) | 85225..85286 | + | 62 | NuclAT_1 | - | Antitoxin |
| - (85225) | 85225..85286 | + | 62 | NuclAT_1 | - | Antitoxin |
| MWM17_RS23880 (85542) | 85542..85616 | - | 75 | Protein_81 | endonuclease | - |
| MWM17_RS23885 (85862) | 85862..86074 | - | 213 | WP_001299730.1 | ANR family transcriptional regulator | - |
| MWM17_RS23890 (86210) | 86210..86770 | - | 561 | WP_000139315.1 | fertility inhibition protein FinO | - |
| MWM17_RS23895 (86873) | 86873..87733 | - | 861 | WP_000704513.1 | alpha/beta hydrolase | - |
| MWM17_RS23900 (87792) | 87792..88538 | - | 747 | WP_109954469.1 | conjugal transfer pilus acetylase TraX | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | tet(A) / dfrA14 / sul2 / aph(3'')-Ib / aph(6)-Id / blaTEM-1B / sitABCD | iroB / iroC / iroD / iroE / iroN / vat / iutA / iucD / iucC / iucB / iucA | 1..171085 | 171085 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 50 a.a. Molecular weight: 5542.67 Da Isoelectric Point: 8.7678
>T40498 WP_001312851.1 NZ_AP025658:c85181-85032 [Escherichia coli]
MTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 150 bp
>T40498 NZ_AP025658:c85181-85032 [Escherichia coli]
ATGACGAAATATGCCCTTATCGGGTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTT
ATGTGAGCTGAATATTCACAGAGGAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
ATGACGAAATATGCCCTTATCGGGTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTT
ATGTGAGCTGAATATTCACAGAGGAAATACAGTGGTGCAGGTAACTCTGGCCTACGAAGCACGGAAGTAA
Antitoxin
Download Length: 62 bp
>AT40498 NZ_AP025658:85225-85286 [Escherichia coli]
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|