Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 2677453..2677678 | Replicon | chromosome |
| Accession | NZ_CP089036 | ||
| Organism | Escherichia coli O157:H7 strain 7386WT | ||
Toxin (Protein)
| Gene name | hokW | Uniprot ID | A0A1Q4PF16 |
| Locus tag | CO544_RS13780 | Protein ID | WP_000813258.1 |
| Coordinates | 2677523..2677678 (+) | Length | 52 a.a. |
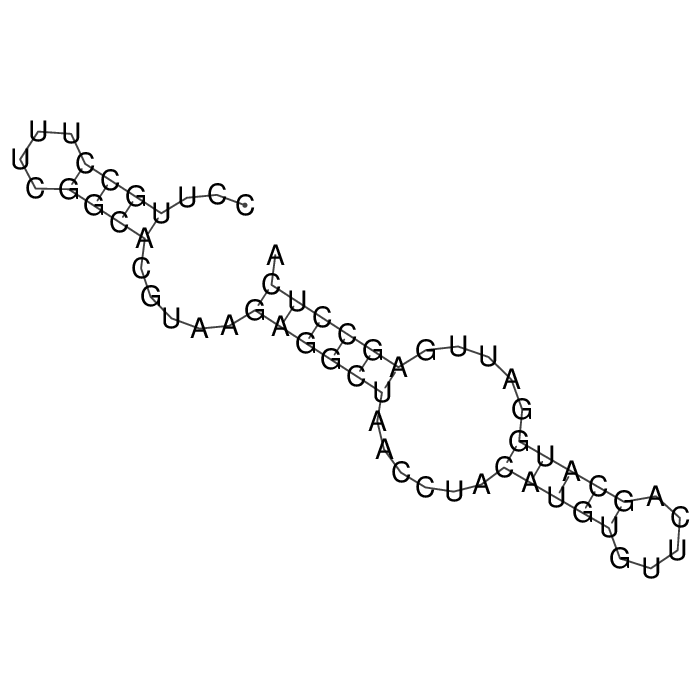
Antitoxin (RNA)
| Gene name | sokW | ||
| Locus tag | - | ||
| Coordinates | 2677453..2677511 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CO544_RS13730 | 2672518..2672760 | + | 243 | WP_000747948.1 | Cro/CI family transcriptional regulator | - |
| CO544_RS13735 | 2672744..2673169 | + | 426 | WP_000693878.1 | toxin YdaT family protein | - |
| CO544_RS13740 | 2673238..2674281 | + | 1044 | WP_001262402.1 | DnaT-like ssDNA-binding domain-containing protein | - |
| CO544_RS13745 | 2674274..2674735 | + | 462 | WP_000139447.1 | replication protein P | - |
| CO544_RS13750 | 2674769..2675485 | + | 717 | WP_000450627.1 | DUF1627 domain-containing protein | - |
| CO544_RS13755 | 2675518..2675799 | + | 282 | WP_000603384.1 | DNA-binding protein | - |
| CO544_RS13760 | 2675796..2676023 | + | 228 | WP_000699809.1 | hypothetical protein | - |
| CO544_RS13765 | 2676016..2676327 | + | 312 | WP_001289673.1 | hypothetical protein | - |
| CO544_RS13770 | 2676455..2676673 | + | 219 | WP_000683609.1 | DUF4014 family protein | - |
| CO544_RS13775 | 2676675..2677232 | + | 558 | WP_000104474.1 | DUF551 domain-containing protein | - |
| - | 2677453..2677511 | - | 59 | - | - | Antitoxin |
| CO544_RS13780 | 2677523..2677678 | + | 156 | WP_000813258.1 | Hok/Gef family protein | Toxin |
| CO544_RS13785 | 2677798..2678142 | + | 345 | WP_000756596.1 | YgiW/YdeI family stress tolerance OB fold protein | - |
| CO544_RS13790 | 2678264..2678536 | + | 273 | WP_000191872.1 | hypothetical protein | - |
| CO544_RS13795 | 2678538..2679587 | + | 1050 | WP_001265229.1 | DUF968 domain-containing protein | - |
| CO544_RS13800 | 2679600..2679905 | + | 306 | WP_001217444.1 | RusA family crossover junction endodeoxyribonuclease | - |
| CO544_RS13805 | 2679968..2680522 | + | 555 | WP_000640035.1 | DUF1133 family protein | - |
| CO544_RS13810 | 2680747..2680944 | + | 198 | WP_000917763.1 | hypothetical protein | - |
| CO544_RS13815 | 2681080..2681793 | + | 714 | WP_000301797.1 | helix-turn-helix domain-containing protein | - |
| CO544_RS13830 | 2682244..2682675 | + | 432 | WP_001302123.1 | tellurite resistance protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Prophage | - | nleG7' | 2609071..2709914 | 100843 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 52 a.a. Molecular weight: 5767.95 Da Isoelectric Point: 7.7942
>T227536 WP_000813258.1 NZ_CP089036:2677523-2677678 [Escherichia coli O157:H7]
MKQQKAMLIALIVICLTVIVTALVTRKDLCEVRIRTGQTEVAVFVDYESEK
MKQQKAMLIALIVICLTVIVTALVTRKDLCEVRIRTGQTEVAVFVDYESEK
Download Length: 156 bp
>T227536 NZ_CP122938:2092816-2092919 [Escherichia coli]
GGCAAGGCAACTAAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATAC
AGGAATCGTGTTCGGTCTCTTTTT
GGCAAGGCAACTAAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATAC
AGGAATCGTGTTCGGTCTCTTTTT
Antitoxin
Download Length: 59 bp
>AT227536 NZ_CP089036:c2677511-2677453 [Escherichia coli O157:H7]
CCTTGCCTTTCGGCACGTAAGAGGCTAACCTACATGTGTTCAGCATGGATTGAGCCTCA
CCTTGCCTTTCGGCACGTAAGAGGCTAACCTACATGTGTTCAGCATGGATTGAGCCTCA
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|