Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 1224058..1224283 | Replicon | chromosome |
| Accession | NZ_CP079891 | ||
| Organism | Escherichia fergusonii strain 5zf15-2-1 | ||
Toxin (Protein)
| Gene name | hokW | Uniprot ID | - |
| Locus tag | KYD87_RS06345 | Protein ID | WP_000813255.1 |
| Coordinates | 1224058..1224213 (-) | Length | 52 a.a. |
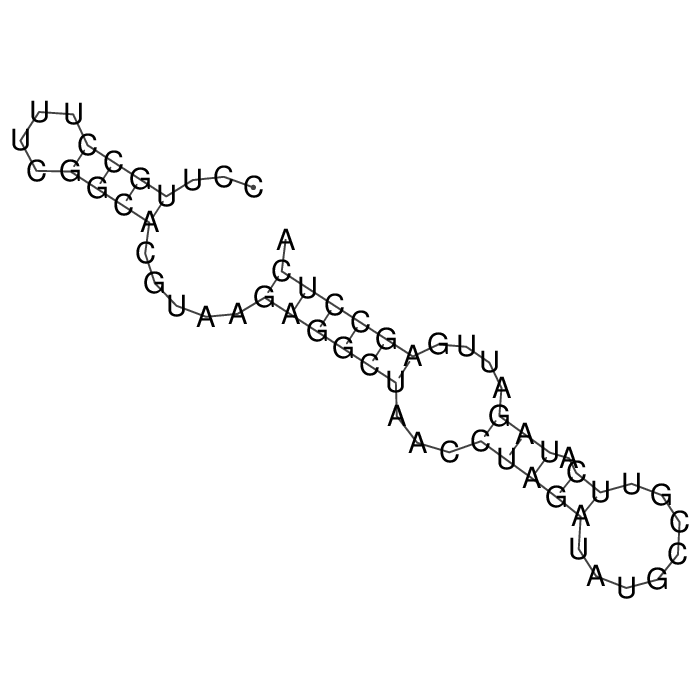
Antitoxin (RNA)
| Gene name | sokW | ||
| Locus tag | - | ||
| Coordinates | 1224225..1224283 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KYD87_RS06285 | 1219160..1219693 | - | 534 | WP_000992100.1 | lysozyme | - |
| KYD87_RS06290 | 1219757..1220107 | - | 351 | WP_000193280.1 | YdfR family protein | - |
| KYD87_RS06295 | 1220112..1220327 | - | 216 | WP_000839572.1 | class II holin family protein | - |
| KYD87_RS06320 | 1221123..1221812 | - | 690 | WP_001064894.1 | bacteriophage antitermination protein Q | - |
| KYD87_RS06325 | 1221809..1222174 | - | 366 | WP_000140004.1 | RusA family crossover junction endodeoxyribonuclease | - |
| KYD87_RS06330 | 1222175..1223230 | - | 1056 | WP_223666277.1 | DUF968 domain-containing protein | - |
| KYD87_RS06335 | 1223232..1223510 | - | 279 | WP_078207408.1 | hypothetical protein | - |
| KYD87_RS06340 | 1223577..1223837 | - | 261 | WP_000687435.1 | hypothetical protein | - |
| KYD87_RS06345 | 1224058..1224213 | - | 156 | WP_000813255.1 | type I toxin-antitoxin system toxin HokD | Toxin |
| - | 1224225..1224283 | + | 59 | - | - | Antitoxin |
| KYD87_RS06350 | 1224456..1224632 | - | 177 | WP_194285216.1 | hypothetical protein | - |
| KYD87_RS06355 | 1224625..1224807 | - | 183 | WP_001224672.1 | hypothetical protein | - |
| KYD87_RS06360 | 1224901..1225257 | - | 357 | WP_039022684.1 | hypothetical protein | - |
| KYD87_RS06365 | 1225315..1225737 | - | 423 | WP_078207410.1 | DUF977 family protein | - |
| KYD87_RS06370 | 1225753..1226523 | - | 771 | WP_039022663.1 | DUF1627 domain-containing protein | - |
| KYD87_RS06375 | 1226556..1227017 | - | 462 | WP_239637900.1 | replication protein P | - |
| KYD87_RS06380 | 1227010..1228050 | - | 1041 | WP_039022662.1 | DnaT-like ssDNA-binding domain-containing protein | - |
| KYD87_RS06385 | 1228022..1228573 | - | 552 | WP_000705377.1 | toxin YdaT family protein | - |
| KYD87_RS06390 | 1228557..1228784 | - | 228 | WP_000476986.1 | Cro/CI family transcriptional regulator | - |
| KYD87_RS06395 | 1228862..1229269 | + | 408 | WP_001003379.1 | helix-turn-helix domain-containing protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Prophage | - | - | 1180941..1239376 | 58435 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 52 a.a. Molecular weight: 5726.85 Da Isoelectric Point: 6.1531
>T210326 WP_000813255.1 NZ_CP079891:c1224213-1224058 [Escherichia fergusonii]
MKQQKAMLIALIVICLTVIVTALVTRKDLCEVRIRTGQTEVAVFTAYESEE
MKQQKAMLIALIVICLTVIVTALVTRKDLCEVRIRTGQTEVAVFTAYESEE
Download Length: 156 bp
>T210326 NZ_CP104500:c2084292-2084185 [Escherichia coli]
ATGACGCTCGCGCAGTTTGCCATGATTTTCTGGCACGACCTGGCAGCACCGATCCTGGCGGGAATTATTACCGCAGCGAT
TGTCGGCTGGTGGCGTAACCGGAAGTAA
ATGACGCTCGCGCAGTTTGCCATGATTTTCTGGCACGACCTGGCAGCACCGATCCTGGCGGGAATTATTACCGCAGCGAT
TGTCGGCTGGTGGCGTAACCGGAAGTAA
Antitoxin
Download Length: 59 bp
>AT210326 NZ_CP079891:1224225-1224283 [Escherichia fergusonii]
CCTTGCCTTTCGGCACGTAAGAGGCTAACCTAGATATGCCGTTCATAGATTGAGCCTCA
CCTTGCCTTTCGGCACGTAAGAGGCTAACCTAGATATGCCGTTCATAGATTGAGCCTCA
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|