Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | sprG-sprF/- |
| Location | 640937..641163 | Replicon | chromosome |
| Accession | NZ_CP077923 | ||
| Organism | Staphylococcus aureus strain 201 | ||
Toxin (Protein)
| Gene name | SprG2 | Uniprot ID | A0A0E0VS87 |
| Locus tag | KU534_RS03370 | Protein ID | WP_000253688.1 |
| Coordinates | 640937..641044 (+) | Length | 36 a.a. |
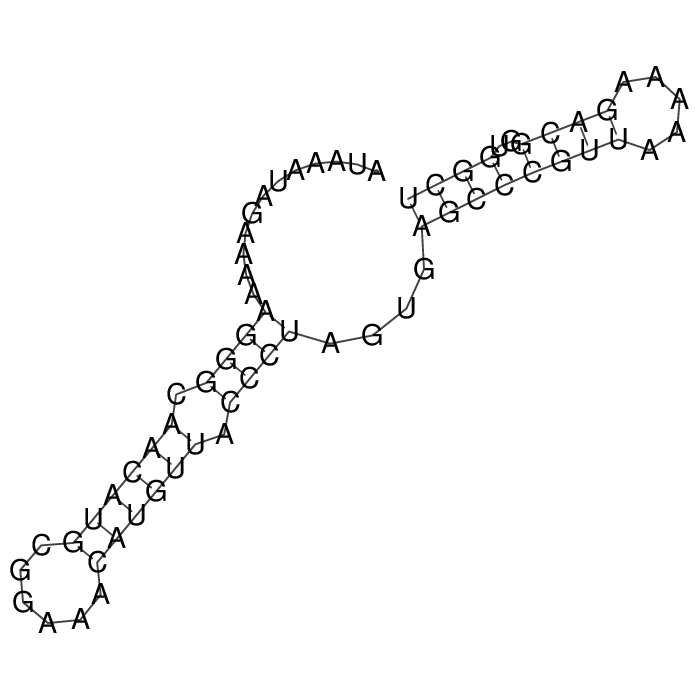
Antitoxin (RNA)
| Gene name | SprF3 | ||
| Locus tag | - | ||
| Coordinates | 641097..641163 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KU534_RS03350 (636198) | 636198..639878 | + | 3681 | WP_000582140.1 | phage tail spike protein | - |
| KU534_RS03355 (639865) | 639865..640017 | + | 153 | WP_001181555.1 | hypothetical protein | - |
| KU534_RS03360 (640064) | 640064..640351 | + | 288 | WP_001040257.1 | hypothetical protein | - |
| KU534_RS03365 (640409) | 640409..640705 | + | 297 | WP_000539688.1 | DUF2951 domain-containing protein | - |
| KU534_RS03370 (640937) | 640937..641044 | + | 108 | WP_000253688.1 | putative holin-like toxin | Toxin |
| - (641097) | 641097..641163 | - | 67 | NuclAT_0 | - | Antitoxin |
| - (641097) | 641097..641163 | - | 67 | NuclAT_0 | - | Antitoxin |
| - (641097) | 641097..641163 | - | 67 | NuclAT_0 | - | Antitoxin |
| - (641097) | 641097..641163 | - | 67 | NuclAT_0 | - | Antitoxin |
| KU534_RS03375 (641088) | 641088..641192 | - | 105 | Protein_660 | hypothetical protein | - |
| KU534_RS03380 (641243) | 641243..641545 | + | 303 | WP_000387656.1 | phage holin | - |
| KU534_RS03385 (641557) | 641557..643011 | + | 1455 | WP_000930259.1 | N-acetylmuramoyl-L-alanine amidase | - |
| KU534_RS03390 (643106) | 643106..643555 | - | 450 | WP_000727649.1 | chemotaxis-inhibiting protein CHIPS | - |
| KU534_RS03395 (644240) | 644240..644590 | + | 351 | WP_000702263.1 | complement inhibitor SCIN-A | - |
| KU534_RS03400 (644643) | 644643..644903 | - | 261 | WP_001791826.1 | hypothetical protein | - |
| KU534_RS03405 (645214) | 645214..645393 | - | 180 | WP_000669789.1 | hypothetical protein | - |
| KU534_RS03410 (645765) | 645765..645962 | - | 198 | WP_000239614.1 | phospholipase | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Prophage | - | groEL / hlb / chp / scn | 593620..644590 | 50970 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 36 a.a. Molecular weight: 3817.78 Da Isoelectric Point: 10.4997
>T208619 WP_000253688.1 NZ_CP077923:640937-641044 [Staphylococcus aureus]
VVSIVDALNLMFSFGMFIVTLLGLVIAIVKLSHKK
VVSIVDALNLMFSFGMFIVTLLGLVIAIVKLSHKK
Download Length: 108 bp
>T208619 NZ_CP103694:189468-189575 [Escherichia coli]
ATGACGTTCGCAGAGCTGGGCATGGCCTTCTGGCATGATTTAGCGGCTCCGGTCATTGCTGGCATTCTTGCCAGTATGAT
CGTGAACTGGCTGAATAAGCGGAAGTAA
ATGACGTTCGCAGAGCTGGGCATGGCCTTCTGGCATGATTTAGCGGCTCCGGTCATTGCTGGCATTCTTGCCAGTATGAT
CGTGAACTGGCTGAATAAGCGGAAGTAA
Antitoxin
Download Length: 67 bp
>AT208619 NZ_CP077923:c641163-641097 [Staphylococcus aureus]
ATAAATAGAAAAAGGGCAACATGCGGAAACATGTTACCCTAGTGAGCCCGTTAAAAAGACGGTGGCT
ATAAATAGAAAAAGGGCAACATGCGGAAACATGTTACCCTAGTGAGCCCGTTAAAAAGACGGTGGCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|