Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 65860..66113 | Replicon | plasmid pS19_141-a |
| Accession | NZ_CP075641 | ||
| Organism | Escherichia coli strain S19-141 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | A0A148HBD8 |
| Locus tag | JFX59_RS24720 | Protein ID | WP_001336447.1 |
| Coordinates | 65860..66009 (-) | Length | 50 a.a. |
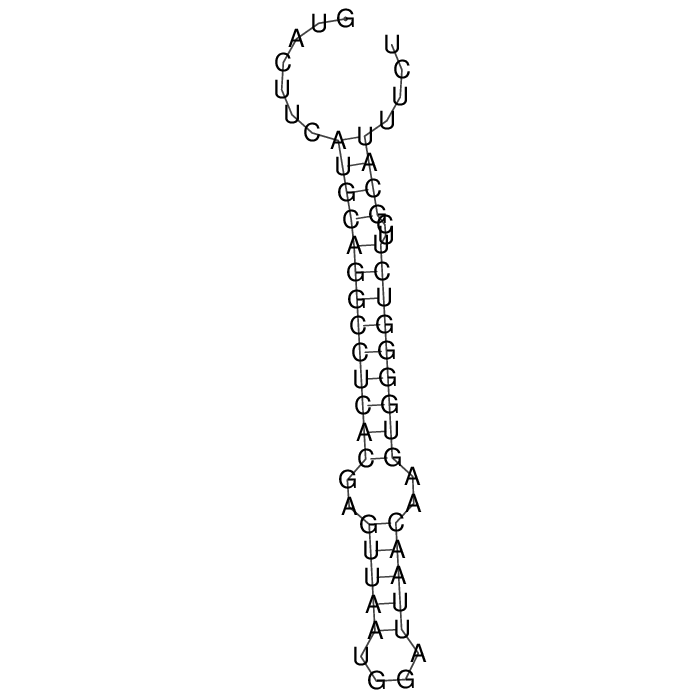
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 66057..66113 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JFX59_RS24695 (62945) | 62945..63184 | - | 240 | WP_000859017.1 | winged helix-turn-helix transcriptional regulator | - |
| JFX59_RS24705 (64160) | 64160..65017 | - | 858 | WP_000130991.1 | incFII family plasmid replication initiator RepA | - |
| JFX59_RS24710 (65010) | 65010..65084 | - | 75 | WP_001365705.1 | RepA leader peptide Tap | - |
| JFX59_RS24715 (65319) | 65319..65576 | - | 258 | WP_000084404.1 | replication regulatory protein RepA | - |
| JFX59_RS24720 (65860) | 65860..66009 | - | 150 | WP_001336447.1 | Hok/Gef family protein | Toxin |
| - (66057) | 66057..66113 | + | 57 | NuclAT_1 | - | Antitoxin |
| - (66057) | 66057..66113 | + | 57 | NuclAT_1 | - | Antitoxin |
| - (66057) | 66057..66113 | + | 57 | NuclAT_1 | - | Antitoxin |
| - (66057) | 66057..66113 | + | 57 | NuclAT_1 | - | Antitoxin |
| JFX59_RS26095 (66288) | 66288..66422 | - | 135 | WP_001058542.1 | hypothetical protein | - |
| JFX59_RS24730 (66773) | 66773..67234 | - | 462 | WP_000760084.1 | thermonuclease family protein | - |
| JFX59_RS24735 (67977) | 67977..68180 | - | 204 | WP_038811942.1 | hypothetical protein | - |
| JFX59_RS24740 (68330) | 68330..68887 | - | 558 | WP_024213676.1 | fertility inhibition protein FinO | - |
| JFX59_RS24745 (68923) | 68923..69291 | - | 369 | WP_000722534.1 | NirD/YgiW/YdeI family stress tolerance protein | - |
| JFX59_RS24750 (69374) | 69374..70120 | - | 747 | WP_024213675.1 | type-F conjugative transfer system pilin acetylase TraX | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | - | hlyD / hlyB / hlyA / hlyC / espP | 1..133041 | 133041 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 50 a.a. Molecular weight: 5572.69 Da Isoelectric Point: 8.7678
>T204073 WP_001336447.1 NZ_CP075641:c66009-65860 [Escherichia coli]
MTKYTLIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MTKYTLIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 150 bp
>T204073 NZ_CP100530:205025-205132 [Escherichia coli]
ATGACGCTCGCAGAACTGGGCATGGCCTTCTGGCATGATTTAGCGGCTCCGGTCATTGCTGGTATTCTTGCCAGTATGAT
CGTGAACTGGCTGAACAAGCGGAAGTAA
ATGACGCTCGCAGAACTGGGCATGGCCTTCTGGCATGATTTAGCGGCTCCGGTCATTGCTGGTATTCTTGCCAGTATGAT
CGTGAACTGGCTGAACAAGCGGAAGTAA
Antitoxin
Download Length: 57 bp
>AT204073 NZ_CP075641:66057-66113 [Escherichia coli]
GTACTTCATGCAGGCCTCACGAGTTAATGGATTAACAAGTGGGGTCTTCGCATTTCT
GTACTTCATGCAGGCCTCACGAGTTAATGGATTAACAAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|