Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | ldrD-symR/Ldr(toxin) |
| Location | 4206649..4206868 | Replicon | chromosome |
| Accession | NZ_CP070951 | ||
| Organism | Escherichia fergusonii strain EF31 | ||
Toxin (Protein)
| Gene name | ldrD | Uniprot ID | S1NWX0 |
| Locus tag | JW961_RS20205 | Protein ID | WP_001295224.1 |
| Coordinates | 4206649..4206756 (-) | Length | 36 a.a. |
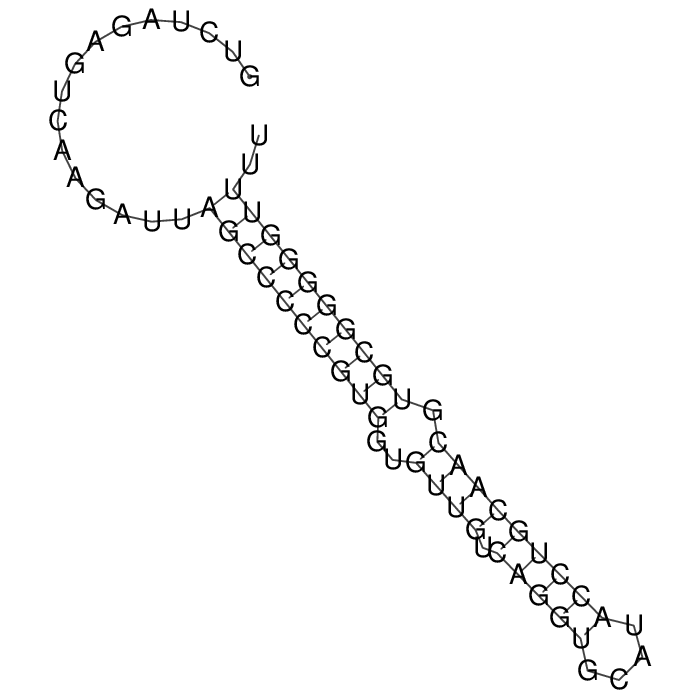
Antitoxin (RNA)
| Gene name | symR | ||
| Locus tag | - | ||
| Coordinates | 4206805..4206868 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JW961_RS20175 (4201698) | 4201698..4201886 | - | 189 | WP_001063310.1 | cellulose biosynthesis protein BcsR | - |
| JW961_RS20180 (4202173) | 4202173..4203732 | + | 1560 | WP_001070267.1 | cellulose biosynthesis c-di-GMP-binding protein BcsE | - |
| JW961_RS20185 (4203729) | 4203729..4203920 | + | 192 | WP_000981122.1 | cellulose biosynthesis protein BcsF | - |
| JW961_RS20190 (4203917) | 4203917..4205596 | + | 1680 | WP_000192007.1 | cellulose biosynthesis protein BcsG | - |
| JW961_RS20195 (4205683) | 4205683..4205790 | - | 108 | WP_002431793.1 | type I toxin-antitoxin system toxin Ldr family protein | - |
| JW961_RS20200 (4206167) | 4206167..4206274 | - | 108 | WP_000170738.1 | type I toxin-antitoxin system toxin Ldr family protein | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_16 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_16 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_16 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_16 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_18 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_18 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_18 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_18 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_20 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_20 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_20 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_20 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_22 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_22 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_22 | - | - |
| - (4206323) | 4206323..4206386 | + | 64 | NuclAT_22 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_11 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_11 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_11 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_11 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_13 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_13 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_13 | - | - |
| - (4206323) | 4206323..4206388 | + | 66 | NuclAT_13 | - | - |
| JW961_RS20205 (4206649) | 4206649..4206756 | - | 108 | WP_001295224.1 | type I toxin-antitoxin system toxin Ldr family protein | Toxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_15 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_15 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_15 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_15 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_17 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_17 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_17 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_17 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_19 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_19 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_19 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_19 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_21 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_21 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_21 | - | Antitoxin |
| - (4206805) | 4206805..4206868 | + | 64 | NuclAT_21 | - | Antitoxin |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_10 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_10 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_10 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_10 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_12 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_12 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_12 | - | - |
| - (4206805) | 4206805..4206870 | + | 66 | NuclAT_12 | - | - |
| JW961_RS20210 (4207193) | 4207193..4208389 | + | 1197 | WP_223662704.1 | methionine gamma-lyase | - |
| JW961_RS20215 (4208639) | 4208639..4209937 | + | 1299 | WP_223662706.1 | amino acid permease | - |
| JW961_RS20220 (4209953) | 4209953..4211164 | - | 1212 | WP_223662708.1 | sigma 54-interacting transcriptional regulator | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 36 a.a. Molecular weight: 3882.70 Da Isoelectric Point: 9.0157
>T194745 WP_001295224.1 NZ_CP070951:c4206756-4206649 [Escherichia fergusonii]
MTLAELGMAFWHDLAAPVIAGILASMIVNWLNKRK
MTLAELGMAFWHDLAAPVIAGILASMIVNWLNKRK
Download Length: 108 bp
>T194745 NZ_CP093957:410551-410646 [Enterococcus faecalis]
ATGTACGAGATTGTCACAAAAATTCTTGTGCCGATTTTTGTCGGGATTGTCCTGAAACTTGTAACCATTTGGTTGGAAAA
ACAGAACGAGGAATAA
ATGTACGAGATTGTCACAAAAATTCTTGTGCCGATTTTTGTCGGGATTGTCCTGAAACTTGTAACCATTTGGTTGGAAAA
ACAGAACGAGGAATAA
Antitoxin
Download Length: 64 bp
>AT194745 NZ_CP070951:4206805-4206868 [Escherichia fergusonii]
GTCTAGAGTCAAGATTAGCCCCCGTGGTGTTGTCAGGTGCATACCTGCAACGTGCGGGGGTTTT
GTCTAGAGTCAAGATTAGCCCCCGTGGTGTTGTCAGGTGCATACCTGCAACGTGCGGGGGTTTT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|